| Experiment ID |
214, 216, 217
|
|---|---|
| Pathogen | |
| Agent Strain |
EPEC B171-8 (serotype O11:NM)
|
| Contains Preferred Model |
No
|
| Reference |
Bieber, D. ., Ramer, S. W., C-y, W. ., Murray, W. J., Tobe, T. ., Fernandez, R. ., & Schoolnik, G. K. (1998). Type IV Pili, Transient Bacterial Aggregates, and Virulence of Enteropathogenic Escherichia coli. Science., 280, 5372. |
| Exposure Route |
oral (with NaHCO3)
|
| Response |
diarrhea
|
| Host type |
human
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # of Doses |
8.00
|
||||||||||||||||||||
| Dose Units |
CFU
|
||||||||||||||||||||
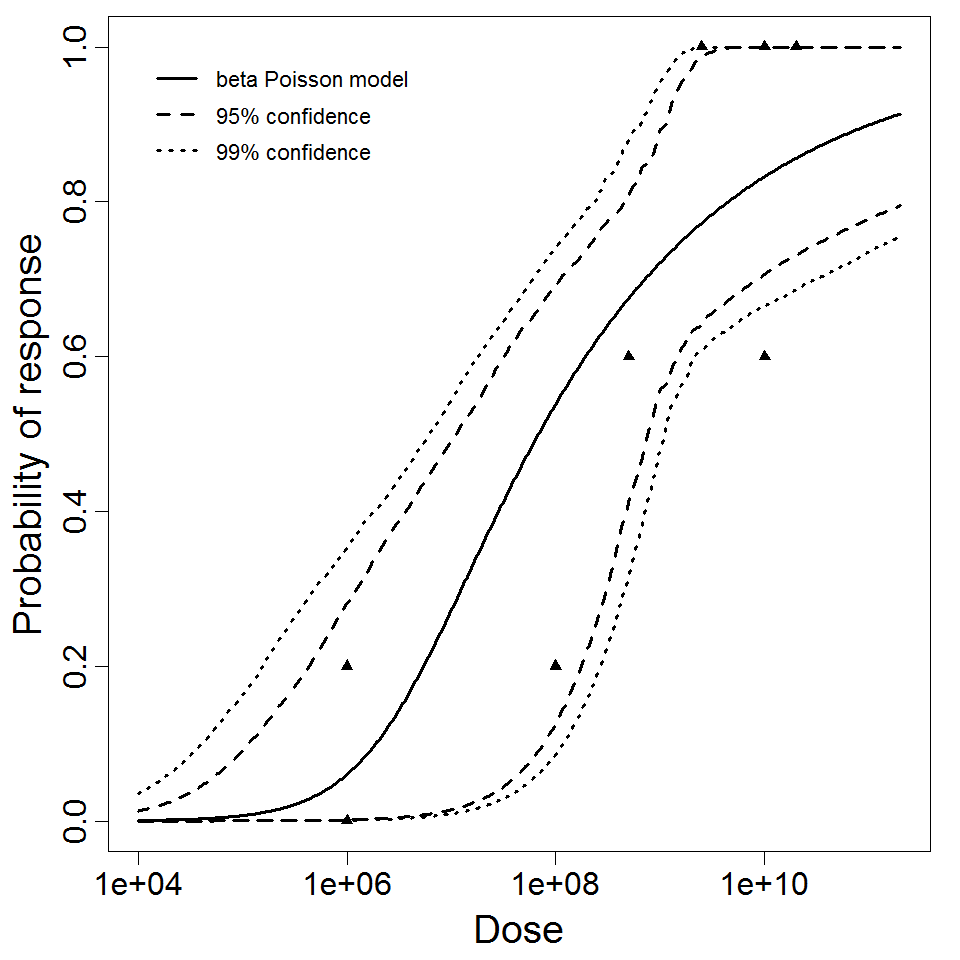
| Μodel |
beta-Poisson
|
||||||||||||||||||||
| a |
2.21E-01
|
||||||||||||||||||||
| N50 |
6.85E+07
|
||||||||||||||||||||
| LD50/ID50 |
6.85E+07
|
||||||||||||||||||||
| Experiment Dataset |
|
Description
|
| ||||||||||||||||||||||
| ||||||||||||||||||||||||||||||