Mice/ Salmonella strain 533 data [1]
| Dose |
Dead |
Survived |
Total |
| 603 |
6 |
36 |
42 |
| 1910 |
3 |
39 |
42 |
| 6030 |
7 |
35 |
42 |
| 19100 |
5 |
42 |
47 |
| 60300 |
6 |
34 |
40 |
| 191000 |
3 |
29 |
32 |
| 603000 |
6 |
20 |
26 |
| 1910000 |
7 |
7 |
14 |
| 6030000 |
7 |
5 |
12 |
| 1.91E+07 |
10 |
2 |
12 |
| 6.03E+07 |
13 |
0 |
13 |
|
Goodness of fit and model selection
| Model |
Deviance |
Δ |
Degrees
of freedom |
χ20.95,1
p-value |
χ20.95,m-k
p-value |
| Exponential |
193 |
146 |
10 |
3.84
0 |
18.3
0 |
| Beta Poisson |
47.5 |
9 |
16.9
3.22e-07 |
| Neither the exponential nor beta-Poisson fits well; beta-Poisson is less bad. |
|
Optimized parameters for the beta-Poisson model, from 10000 bootstrap iterations
| Parameter |
MLE estimate |
Percentiles |
| 0.5% |
2.5% |
5% |
95% |
97.5% |
99.5% |
| α |
6.21E-02 |
3.53E-02 |
4.06E-02 |
4.32E-02 |
1.09E-01 |
1.25E-01 |
1.80E-01 |
| N50 |
3.46E+07 |
9.76E+05 |
1.69E+06 |
2.40E+06 |
9.41E+08 |
2.13E+09 |
1.34E+10 |
|
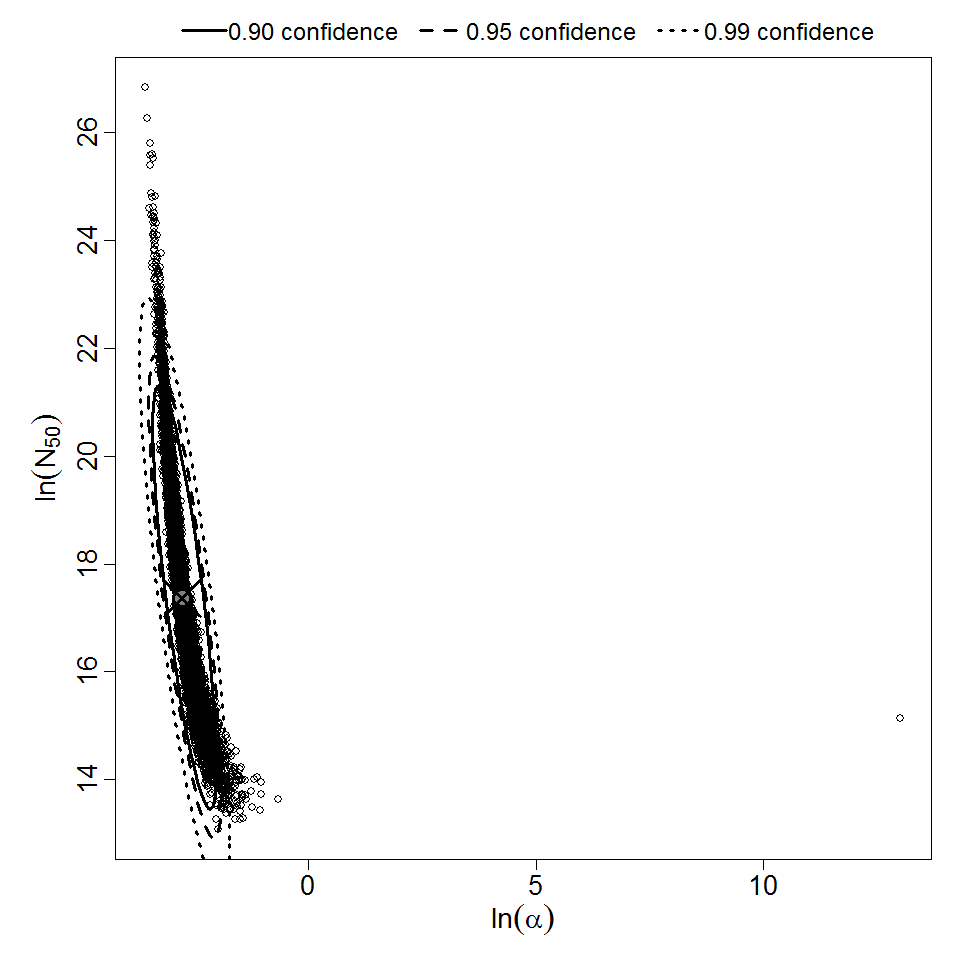
Parameter scatter plot for beta Poisson model ellipses signify the 0.9, 0.95 and 0.99 confidence of the parameters.
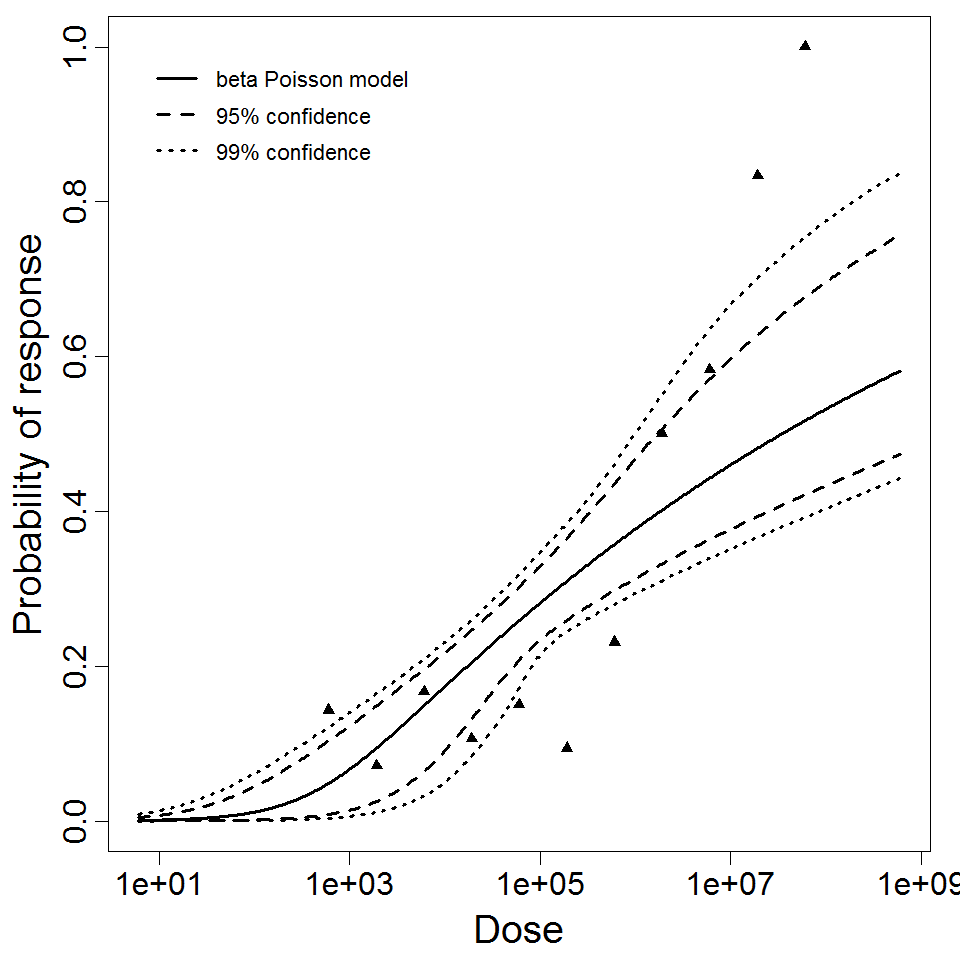
beta Poisson model plot, with confidence bounds around optimized model
References


 QMRA
QMRA