General Overview
Listeria monocytogenes is a Gram-positive rod-shaped bacterium. It is the causative agent of listeriosis, a serious infection caused by eating food contaminated with the bacteria. The disease generally affects older adults, pregnant women, newborns, and adults with weakened immune systems. However, rarely, persons without these risk factors can also be affected, according to the CDC and Todar.
The overt form of the disease has mortality greater than 25 percent. The two main clinical manifestations are sepsis and meningitis. Meningitis is often complicated by encephalitis, a pathology that is unusual for bacterial infections, according to Todar .
Summary Data
Czuprynski et al. studied susceptibility of C57BL/6 mice and A/J mice to Listeria monocytogenes (strain Scott A) inoculating them intragastrically. Similarly, Golnazarian et al. also compared infectious dose in normal and compromised C57BL/6J mice with pathogens( strain F5817) via oral route.
| |||||||||||||||||||||||||||||||||||||||||||||||||||||||
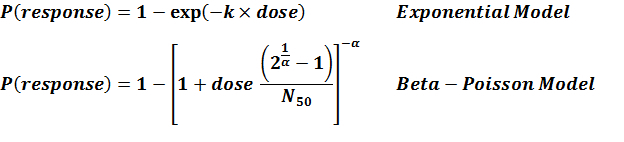
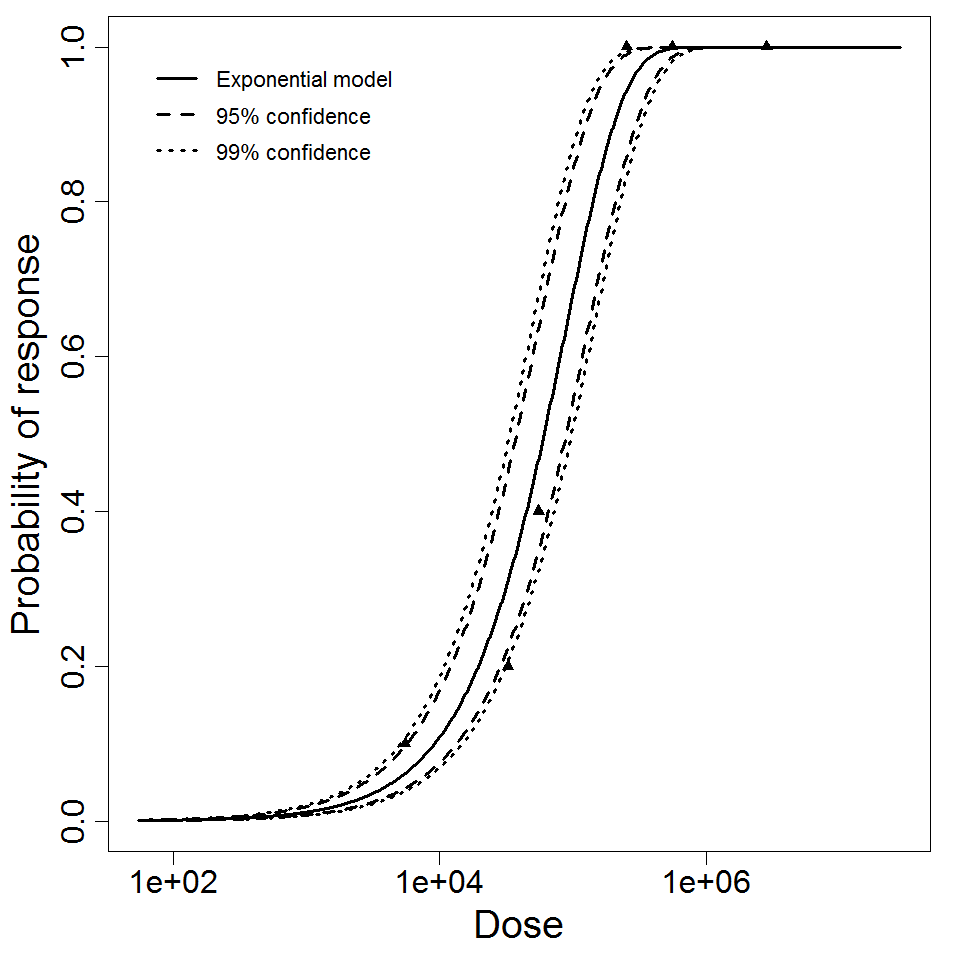
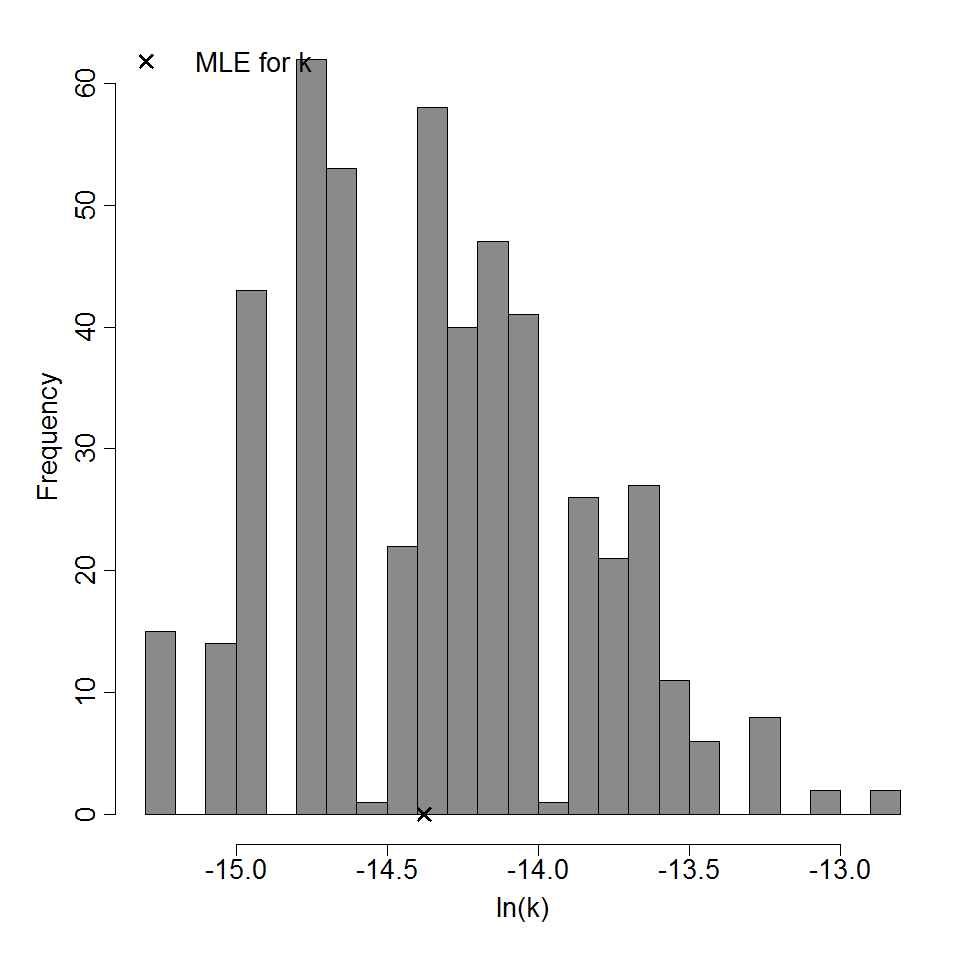
Optimization Output for experiment 291
|
| ||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||
Optimization Output for experiment 293
|
| ||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||
Optimization Output for experiment 294
|
| ||||||||||||||||||||||
| ||||||||||||||||||||||||||||||
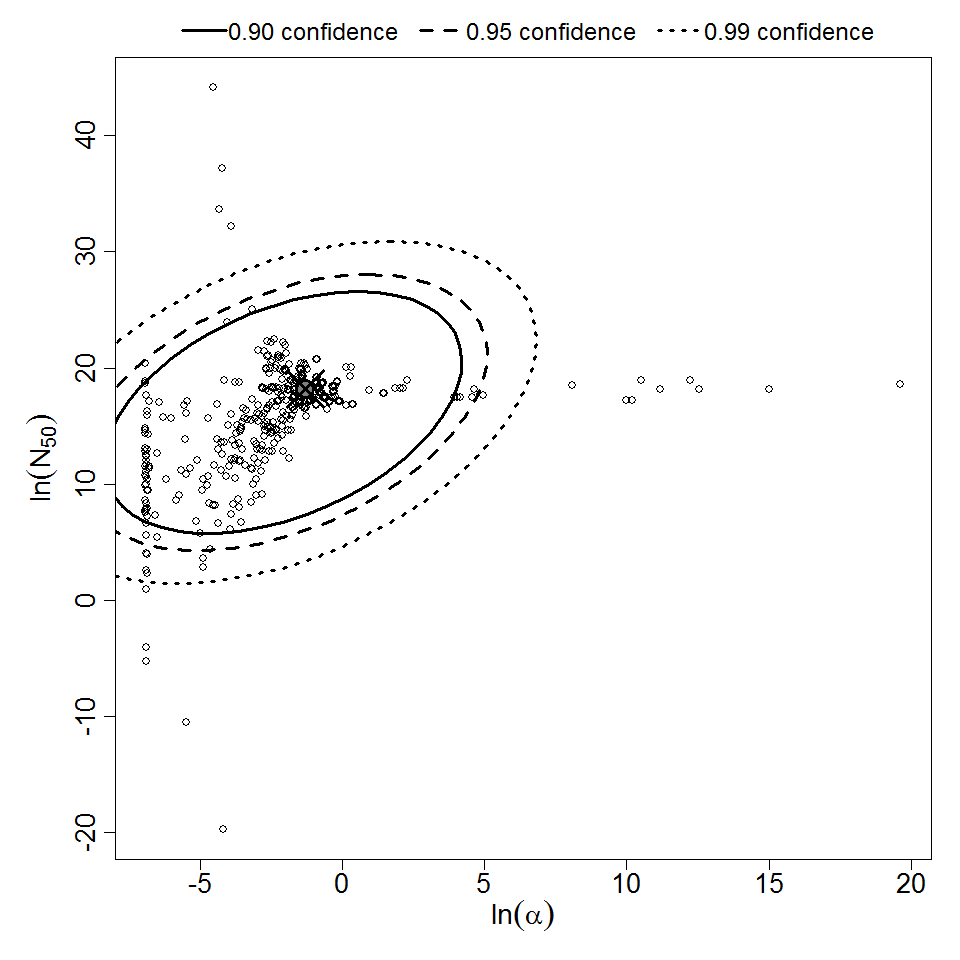
Parameter scatter plot for beta Poisson model ellipses signify the 0.9, 0.95 and 0.99 confidence of the parameters.
| ID | Exposure Route | # of Doses | Agent Strain | Dose Units | Host type | Μodel | LD50/ID50 | Optimized parameters | Response type | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| 292 | oral | 6.00 | F5817 | CFU | beta-Poisson | a = 0.253 N50 = 277 | infection |
| Model | Deviance | Δ | Degrees of freedom |
χ20.95,1 p-value |
χ20.95,m-k p-value |
| Exponential | 9.88 | 6.48 | 5 | 3.84 0.011 |
11.1 0.0788 |
| beta Poisson | 3.4 | 4 | 9.49 0.494 |
||
| beta-Poisson fits better than exponential; can not reject good fit for beta-Poisson | |||||
| Parameter | MLE Estimate | 0.5% | 2.5% | 5% | 95% | 97.5% | 99.5% |
| α | 6.95E-01 | 2.69E-01 | 3.39E-01 | 3.78E-01 | 2.56E+0 | 2.28E+01 | 1.18E+03 |
| N50 | 3.39E+03 | 3.58E+01 | 2.47E+02 | 4.67E+02 | 1.09E+04 | 1.26E+04 | 1.85E+04 |