| ID | Exposure Route | # of Doses | Agent Strain | Dose Units | Host type | Μodel | LD50/ID50 | Optimized parameters | Response type | Reference |
|---|---|---|---|---|---|---|---|---|---|---|
| 281 | injected in eyelids | 6.00 | ATCC 19660 | CFU | Swiss webster mice (5day old) | exponential | 8.13E+03 | k = 8.52E-05 | death |
Hazlett, L. D., Rosen, D. D., & Berk, R. S. (1978). Age-Related Susceptibility to Pseudomonas aeruginosa Ocular Infections in Mice. Infection and Immunity, 20, 1. |
| 281,282 (pooled) | injected in eyelids | 12.00 | ATCC 19660 | CFU | Swiss webster mice (5day old) | exponential | 6.61E+03 | k = 1.05E-04 | death |
Hazlett, L. D., Rosen, D. D., & Berk, R. S. (1978). Age-Related Susceptibility to Pseudomonas aeruginosa Ocular Infections in Mice. Infection and Immunity, 20, 1. |
| 282 | injected in eyelids | 6.00 | ATCC 19660 | CFU | Swiss webster mice (5day old) | exponential | 4.98E+03 | k = 1.39E-04 | death(after day 21) |
Hazlett, L. D., Rosen, D. D., & Berk, R. S. (1978). Age-Related Susceptibility to Pseudomonas aeruginosa Ocular Infections in Mice. Infection and Immunity, 20, 1. |
| 283 | injected in eyelids | 6.00 | ATCC 19660 | CFU | Swiss webster mice(10day old) | beta-Poisson | 1.93E+04 | a = 6.73E-01 N50 = 1.93E+04 | death(after day 1) |
Hazlett, L. D., Rosen, D. D., & Berk, R. S. (1978). Age-Related Susceptibility to Pseudomonas aeruginosa Ocular Infections in Mice. Infection and Immunity, 20, 1. |
| 283,284 | injected in eyelids | 12.00 | ATCC 19660 | CFU | Swiss webster mice(10day old) | beta-Poisson | 1.48E+04 | a = 6.01E-01 N50 = 1.48E+04 | death |
Hazlett, L. D., Rosen, D. D., & Berk, R. S. (1978). Age-Related Susceptibility to Pseudomonas aeruginosa Ocular Infections in Mice. Infection and Immunity, 20, 1. |
| 284 | injected in eyelids | 6.00 | ATCC 19660 | CFU | Swiss webster mice(10day old) | beta-Poisson | 1.13E+04 | a = 5.49E-01 N50 = 1.13E+04 | death(after day 2-21) |
Hazlett, L. D., Rosen, D. D., & Berk, R. S. (1978). Age-Related Susceptibility to Pseudomonas aeruginosa Ocular Infections in Mice. Infection and Immunity, 20, 1. |
| 286 | contact lens | 5.00 | ATCC 19660 | CFU | Swiss webster mice | exponential | 7.88E+06 | k = 8.8E-08 | infection (Keratitis) |
Hazlett, L. D., Rosen, D. D., & Berk, R. S. (1978). Age-Related Susceptibility to Pseudomonas aeruginosa Ocular Infections in Mice. Infection and Immunity, 20, 1. |
| 297 | contact lens | 5.00 | CFU | white rabbit | beta-Poisson | 6.57E+03 | a = 3.55E-01 N50 = 6.57E+03 | corneal ulceration |
Lawin-Brüssel, C. A., Refojo, M. F., Leong, F. L., Hanninen, L. ., & Kenyon, K. R. (1993). Effect of Pseudomonas aeruginosa concentration in experimental contact lens-related microbial keratitis. Cornea, 12, 1. |
|
| 297 & 298 | contact lens | 10.00 | CFU | white rabbit | beta-Poisson | 1.85E+04 | a = 1.9E-01 N50 = 1.85E+04 | corneal ulceration |
Lawin-Brüssel, C. A., Refojo, M. F., Leong, F. L., Hanninen, L. ., & Kenyon, K. R. (1993). Effect of Pseudomonas aeruginosa concentration in experimental contact lens-related microbial keratitis. Cornea, 12, 1. |
|
| 298 | contact lens | 5.00 | CFU | white rabbit | beta-Poisson | 1.52E+05 | a = 1.09E-01 N50 = 1.52E+05 | Severe stromal ulceration |
Lawin-Brüssel, C. A., Refojo, M. F., Leong, F. L., Hanninen, L. ., & Kenyon, K. R. (1993). Effect of Pseudomonas aeruginosa concentration in experimental contact lens-related microbial keratitis. Cornea, 12, 1. |
|
| Ojielo2003 | intratracheal | 7.00 | PA01 | CFU | C57BL/6 mice | exponential | k = 3.22E-7 | death |
Ojielo, C. I., Cooke, K. ., Mancuso, P. ., Standiford, T. J., Olkiewicz, K. M., Clouthier, S. ., … Moore, B. B. (2003). Defective Phagocytosis and Clearance of <i>Pseudomonas aeruginosa</i> in the Lung Following Bone Marrow Transplantation. The Journal of Immunology, 171(8), 4416-4424. Retrieved from https://journals.aai.org/jimmunol/article/171/8/4416/35775/Defective-Phagocytosis-and-Clearance-of (Original work published) |
|
|
||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||
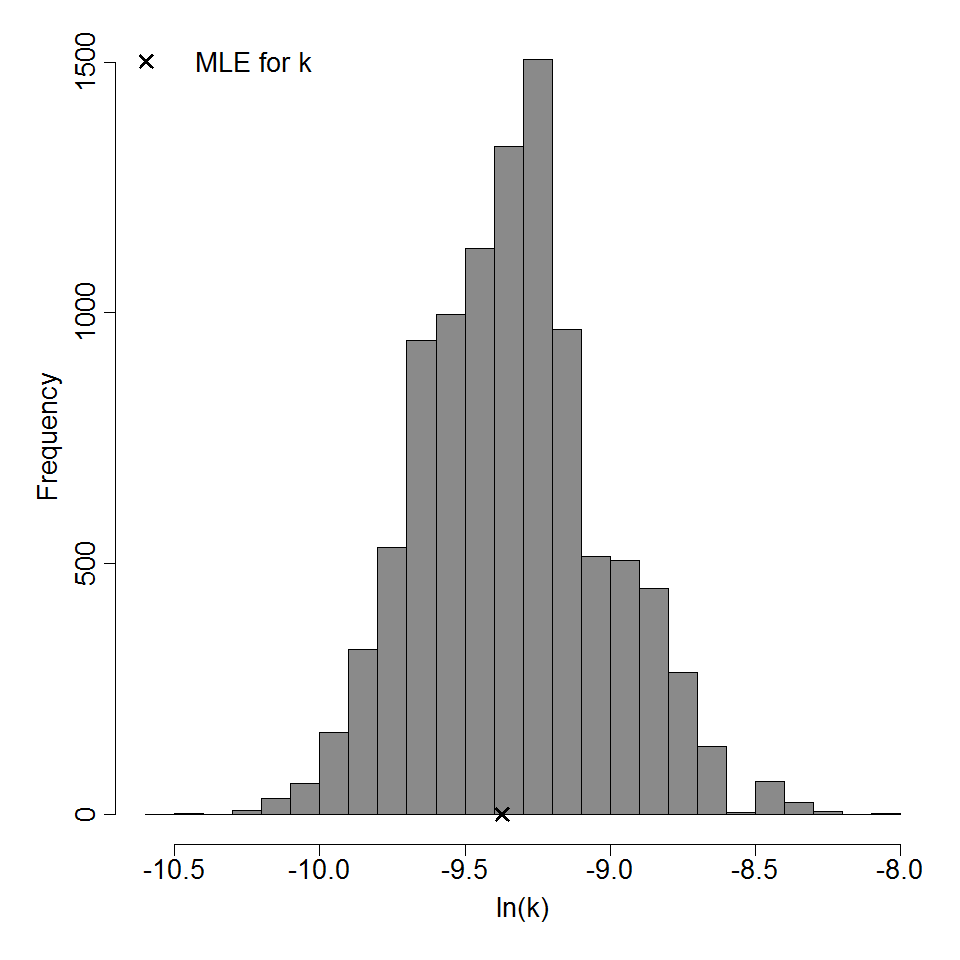
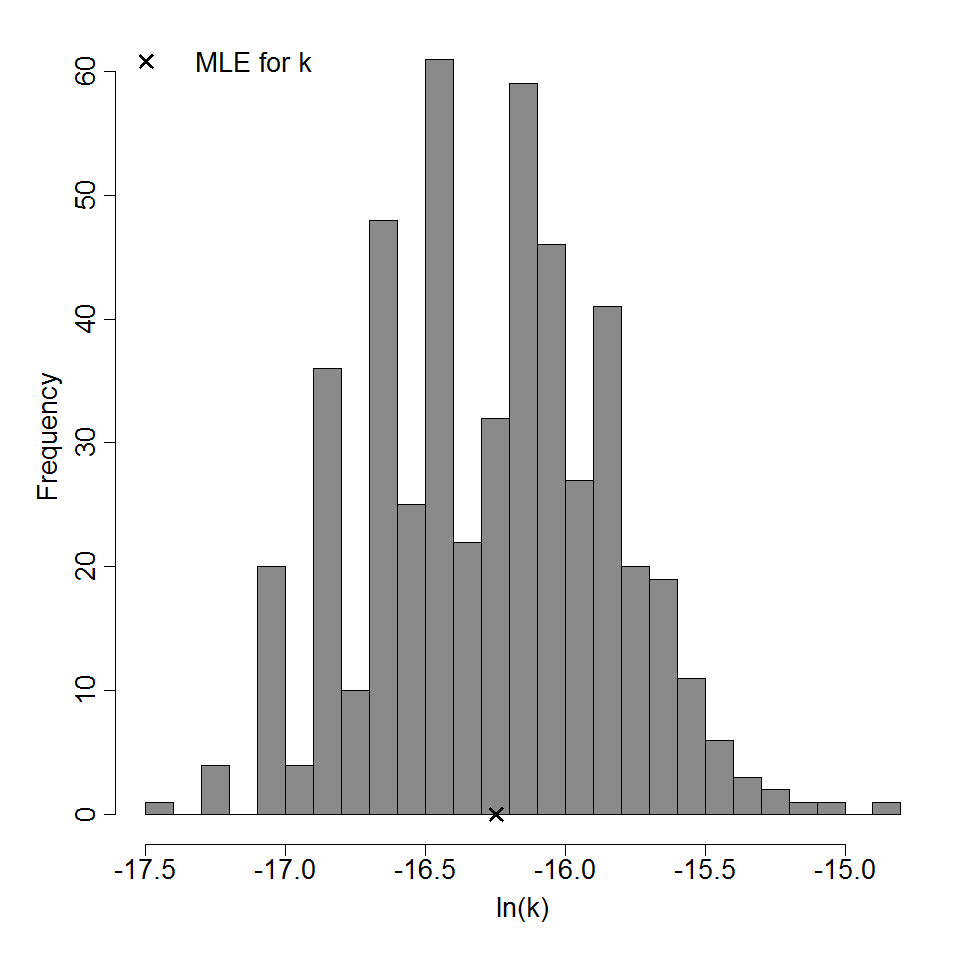
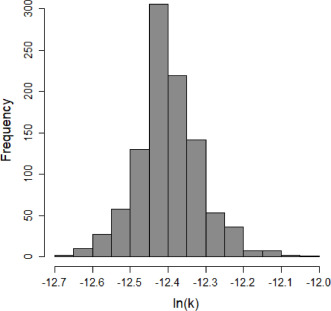
Parameter histogram for exponential model (uncertainty of the parameter)
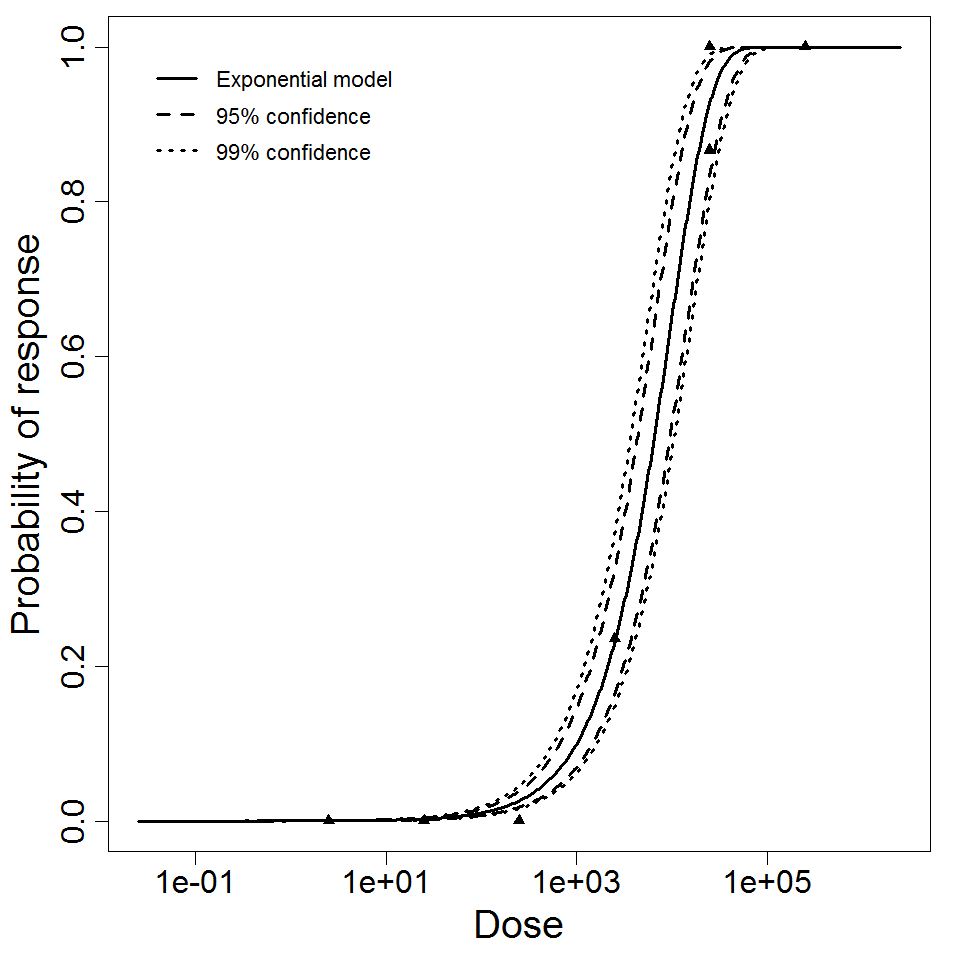
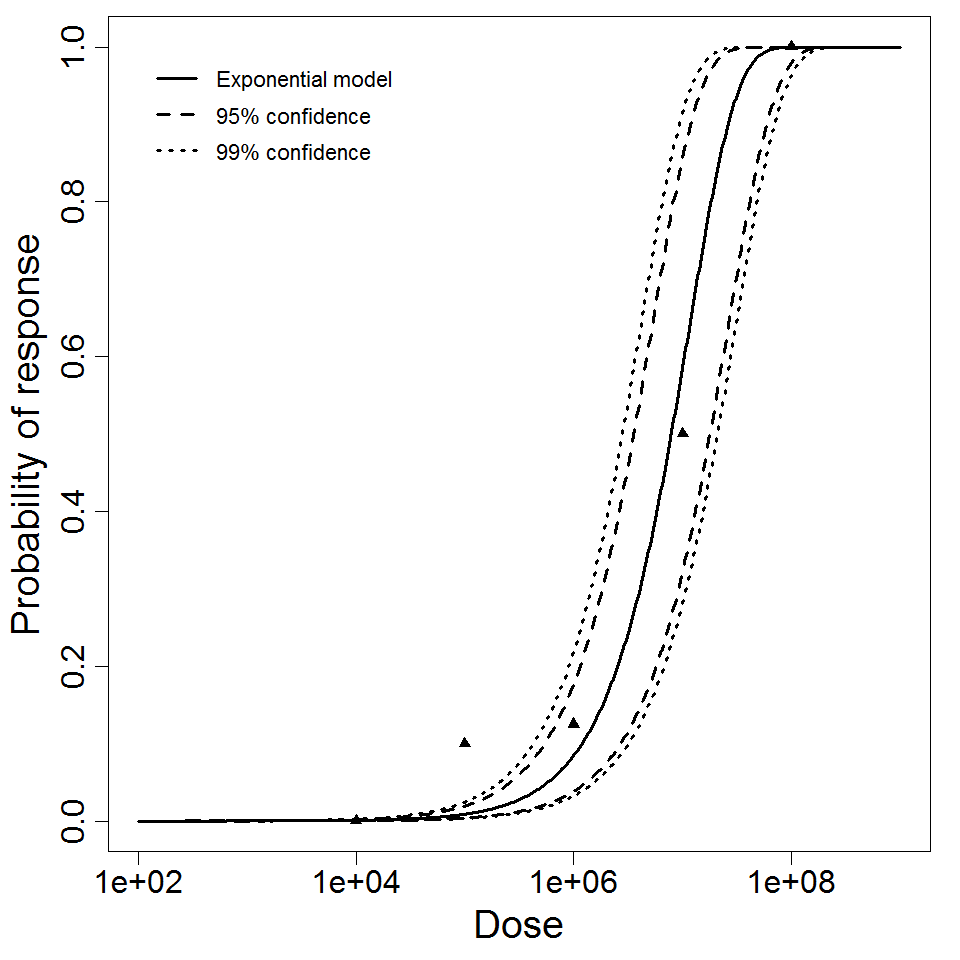
Exponential model plot, with confidence bounds around optimized model
|
|
||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||
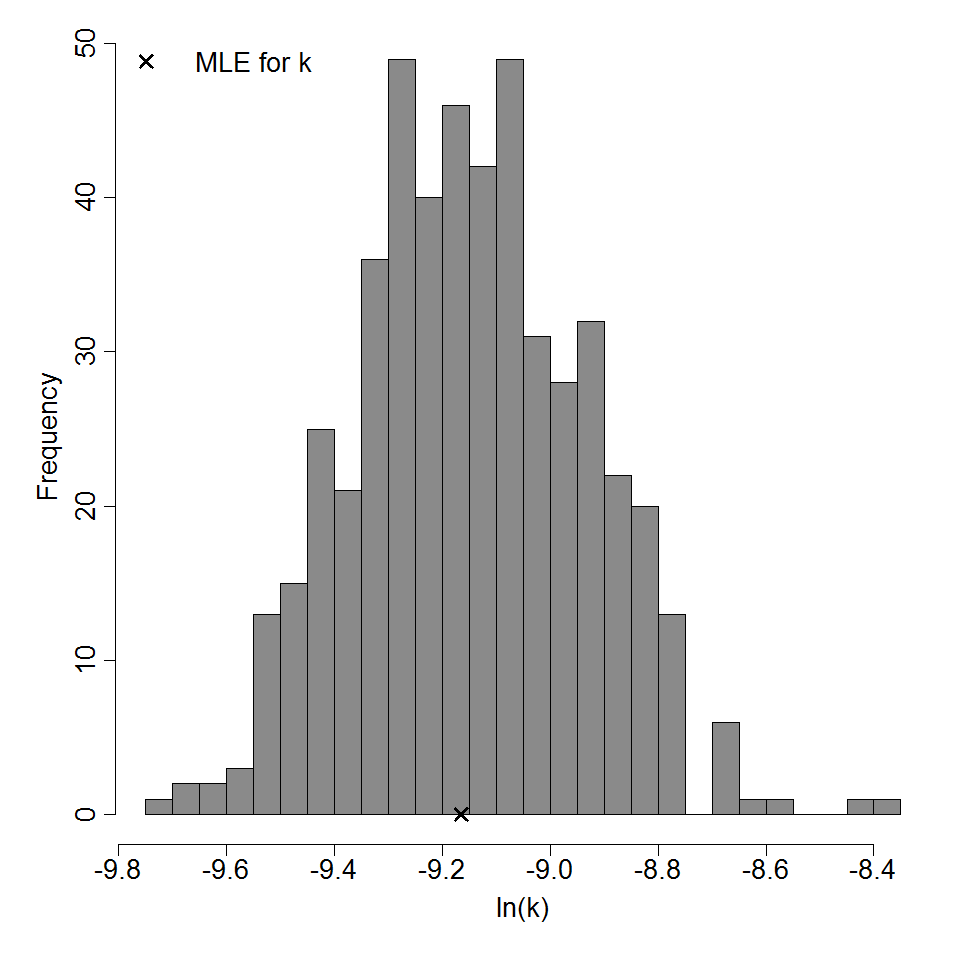
Parameter histogram for exponential model (uncertainty of the parameter)
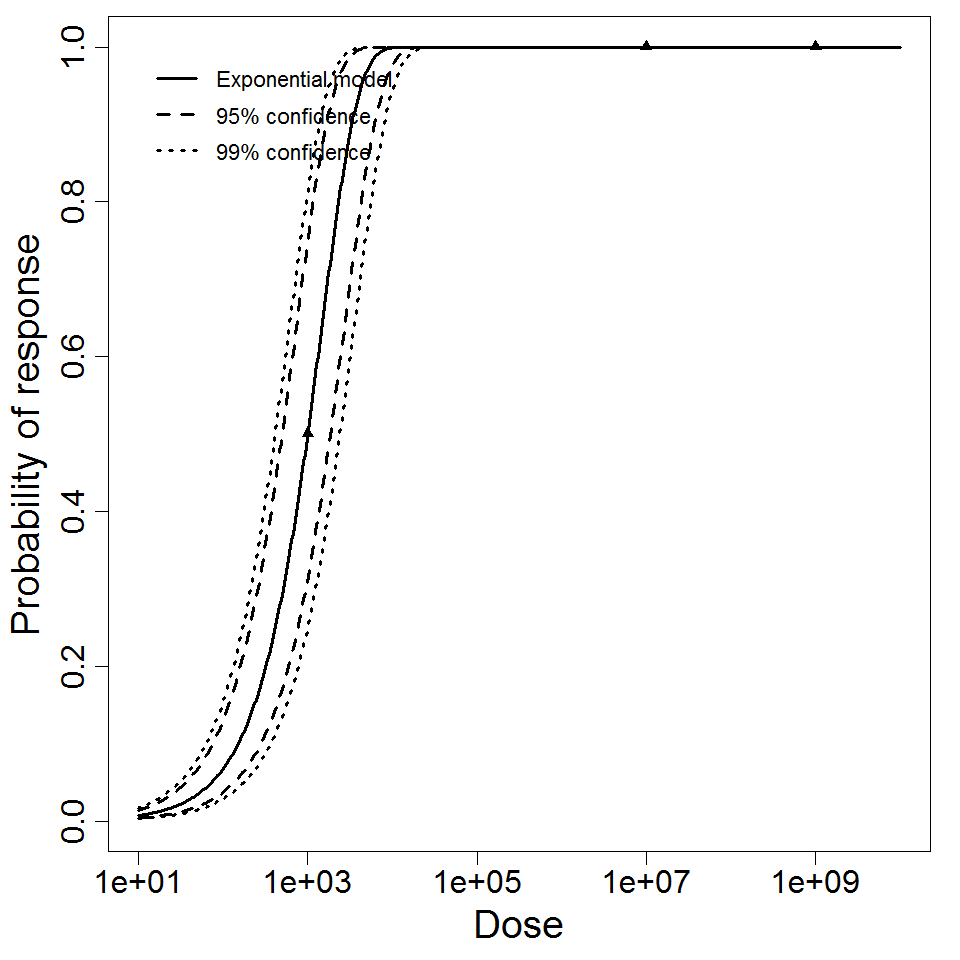
Exponential model plot, with confidence bounds around optimized model
|
|
||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||
Parameter histogram for exponential model (uncertainty of the parameter)
Exponential model plot, with confidence bounds around optimized model
|
|
||||||||||||||||||||||
|
||||||||||||||||||||||||||||||
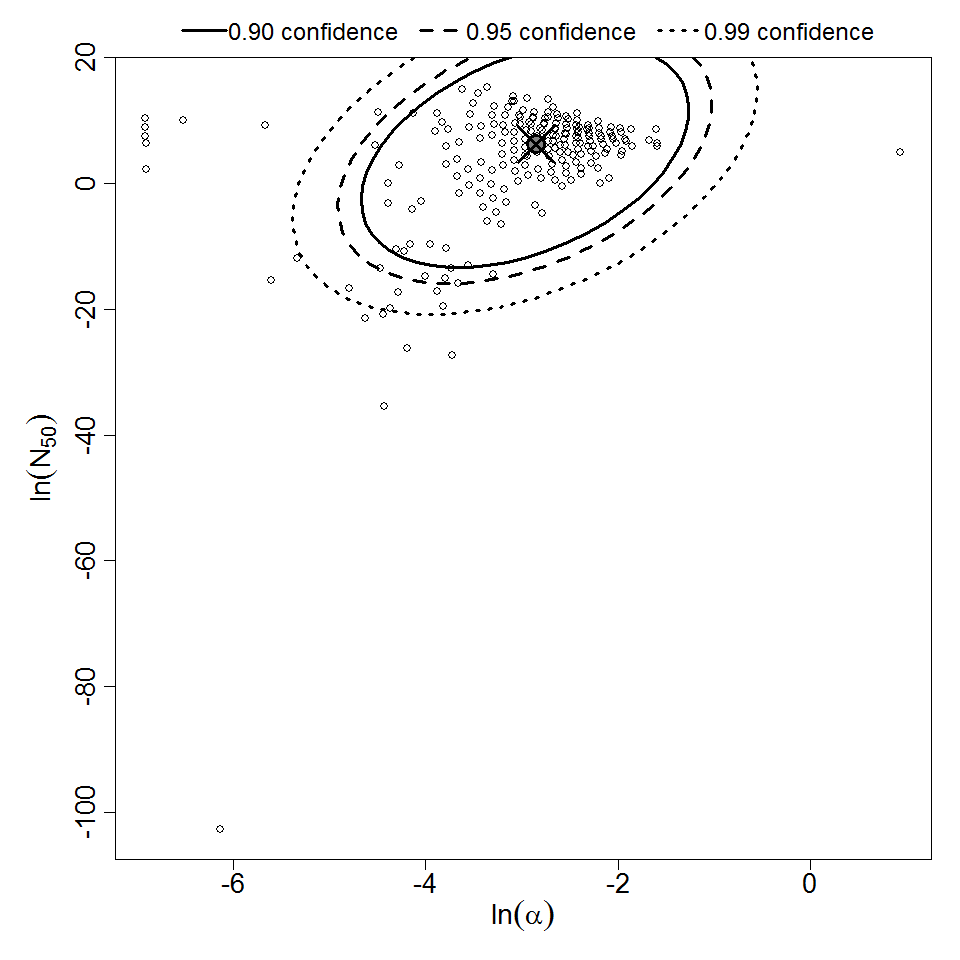
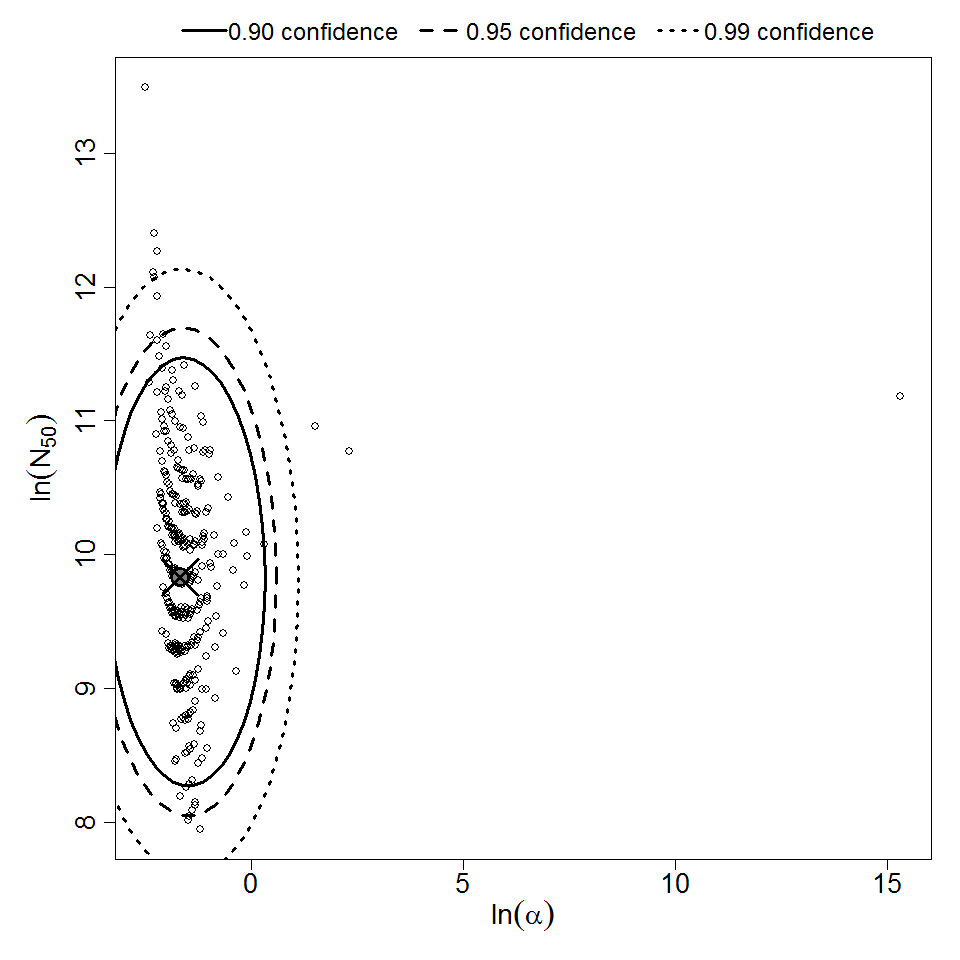
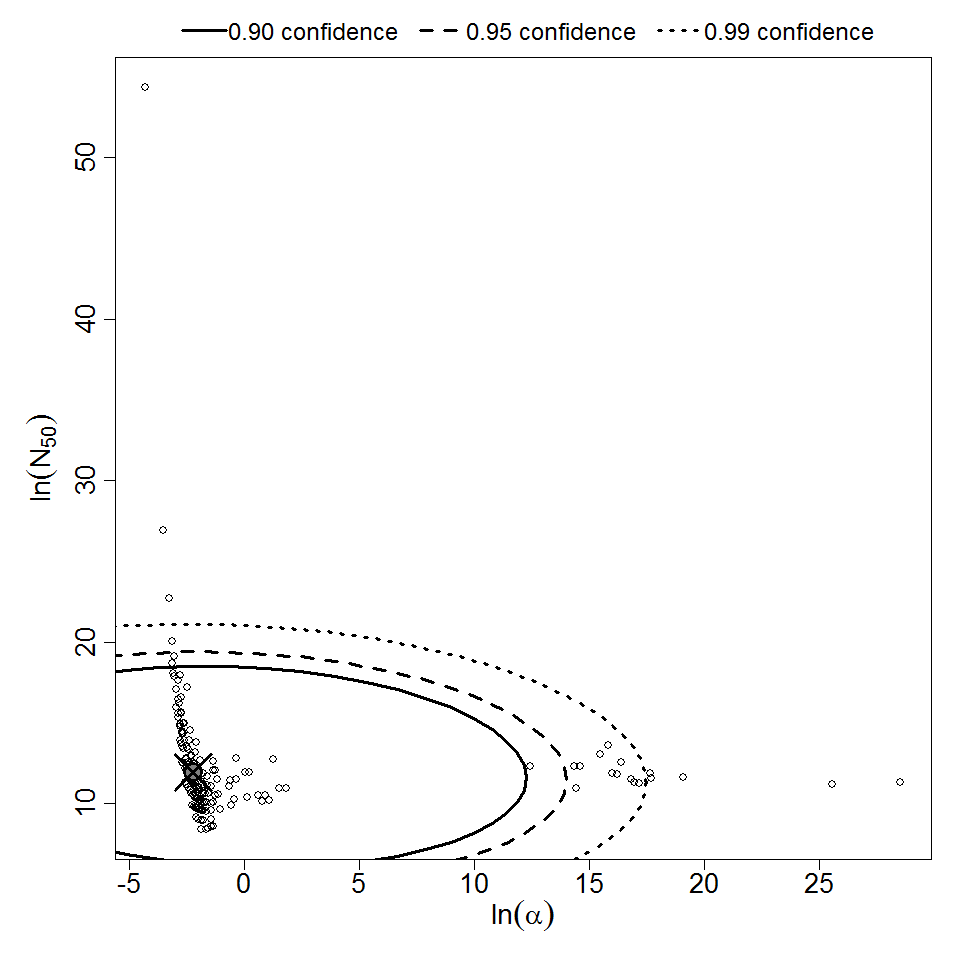
Parameter scatter plot for beta Poisson model ellipses signify the 0.9, 0.95 and 0.99 confidence of the parameters.
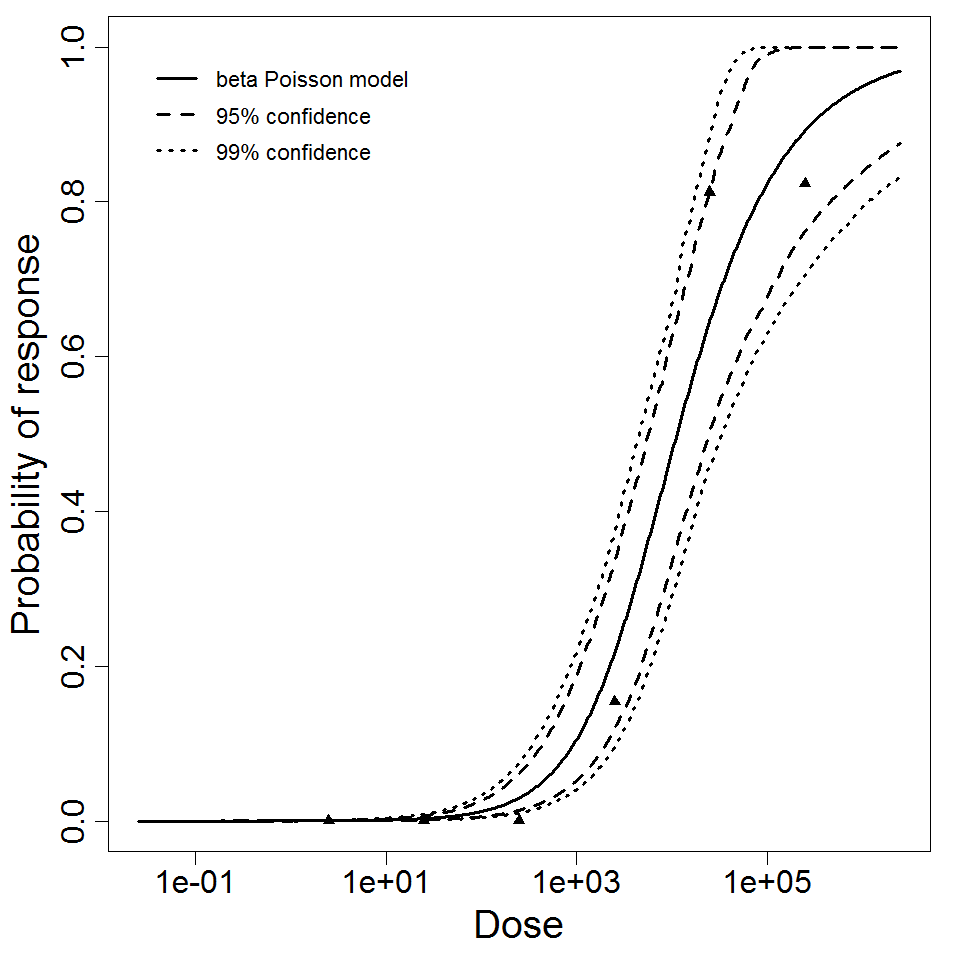
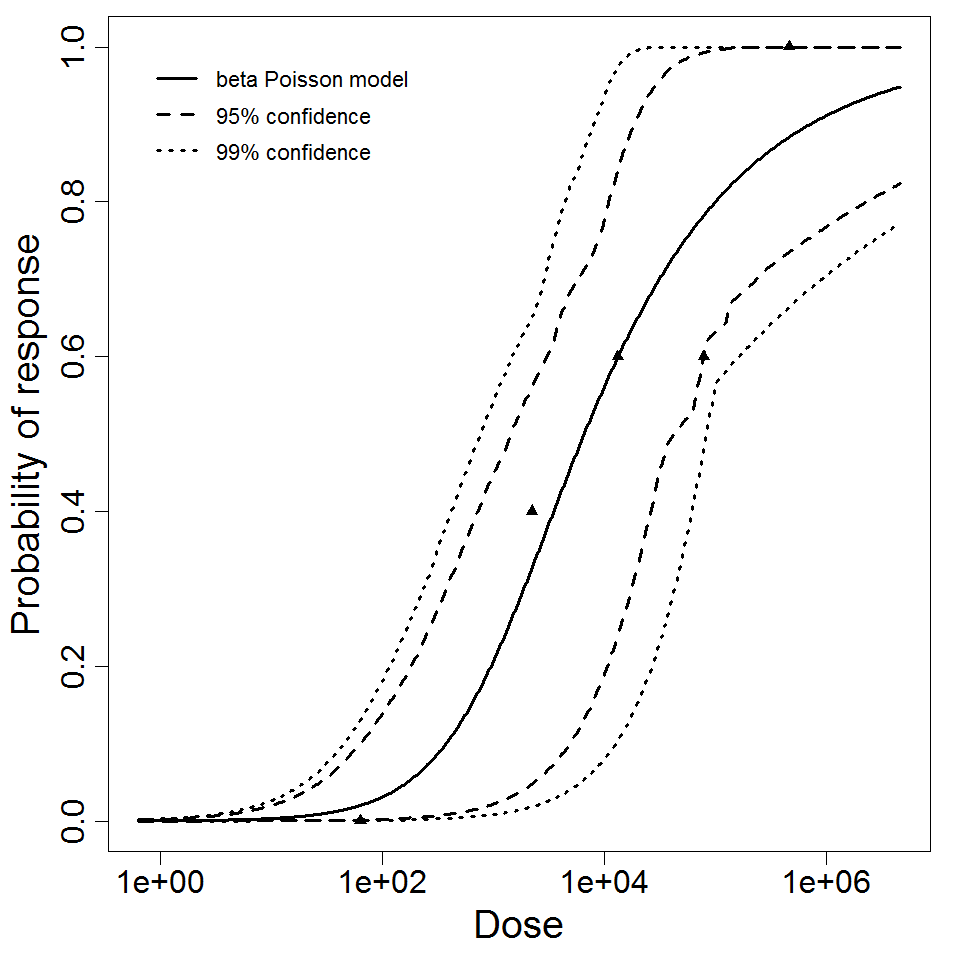
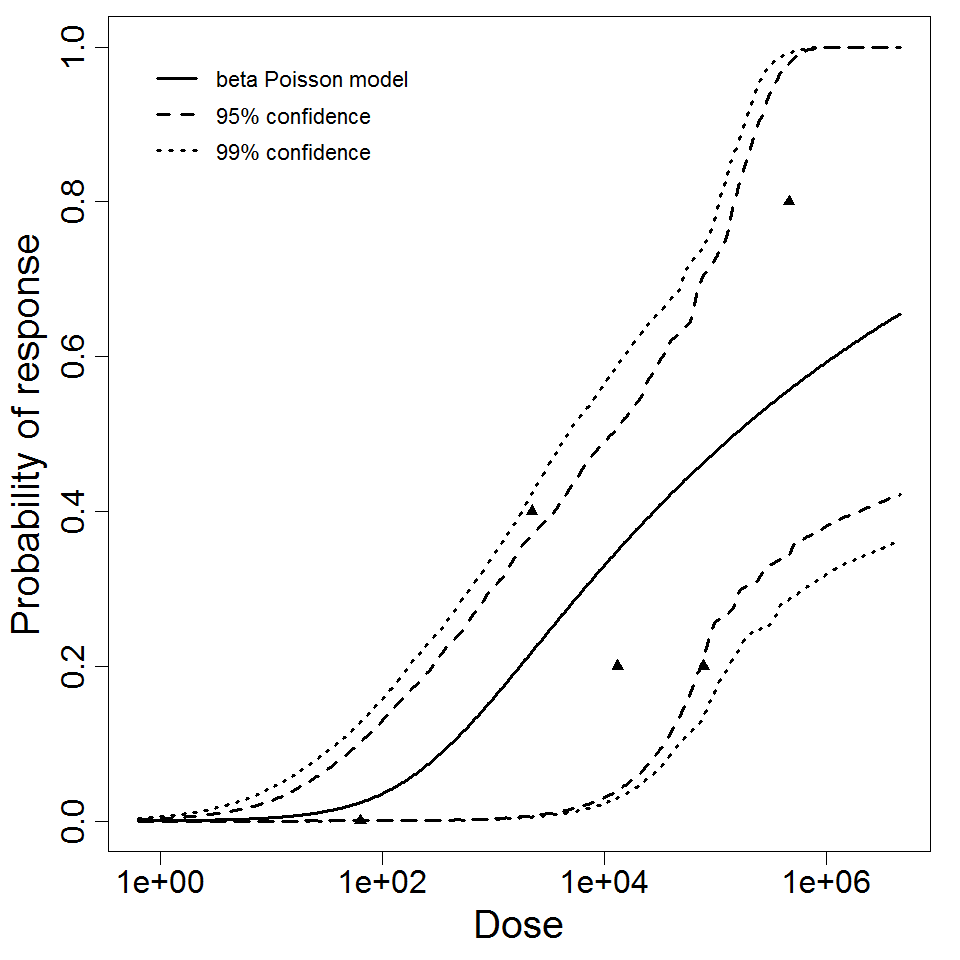
beta Poisson model plot, with confidence bounds around optimized model
|
||||||||||||||||||||||
|
||||||||||||||||||||||||||||||
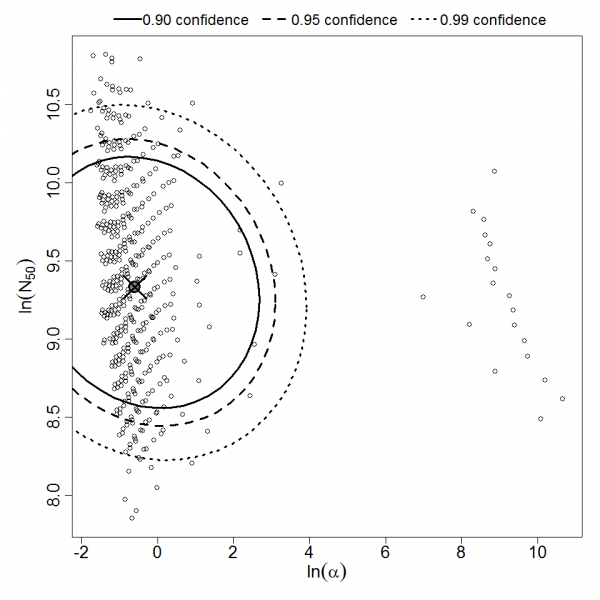
Parameter scatter plot for beta Poisson model ellipses signify the 0.9, 0.95 and 0.99 confidence of the parameters.
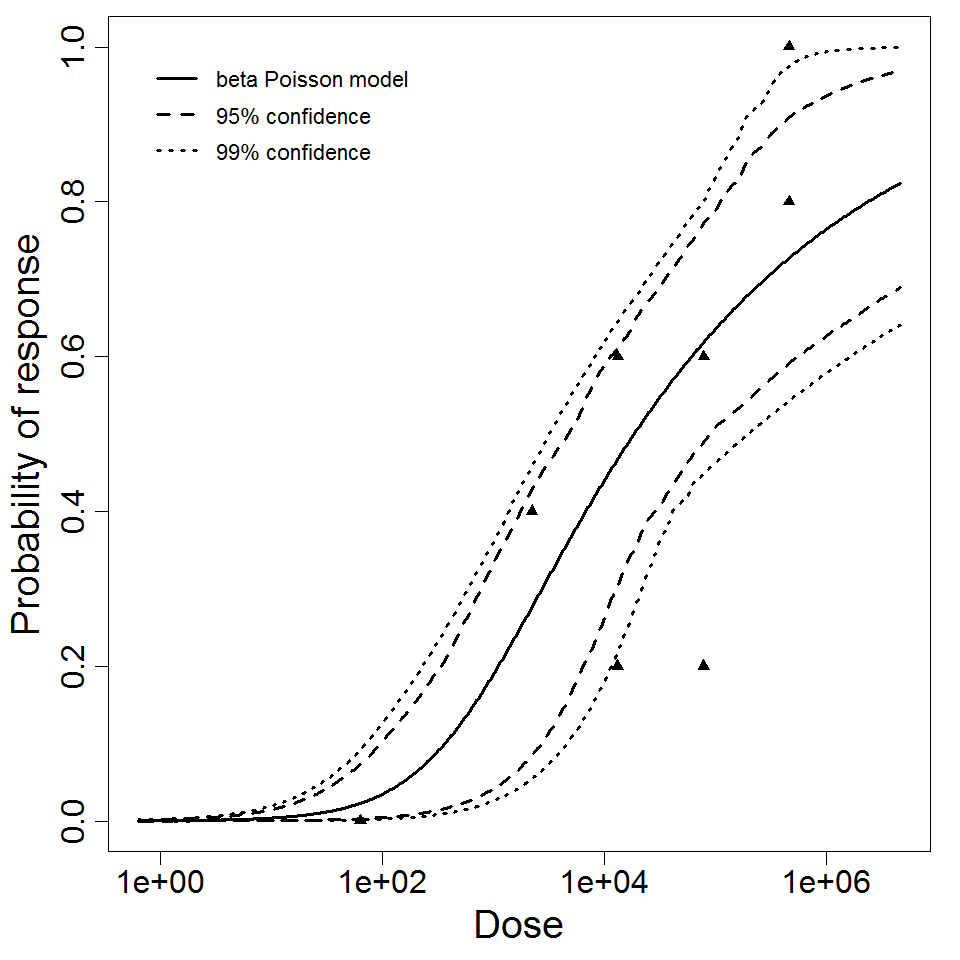
beta Poisson model plot, with confidence bounds around optimized model
|
|
||||||||||||||||||||||
|
||||||||||||||||||||||||||||||
Parameter scatter plot for beta Poisson model ellipses signify the 0.9, 0.95 and 0.99 confidence of the parameters.
beta Poisson model plot, with confidence bounds around optimized model
|
|
||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||
Parameter histogram for exponential model (uncertainty of the parameter)
Exponential model plot, with confidence bounds around optimized model
|
|
||||||||||||||||||||||
|
||||||||||||||||||||||||||||||
Parameter histogram for exponential model (uncertainty of the parameter)
Exponential model plot, with confidence bounds around optimized model
|
|
||||||||||||||||||||||
|
||||||||||||||||||||||||||||||
Parameter histogram for exponential model (uncertainty of the parameter)
Exponential model plot, with confidence bounds around optimized model
|
|
||||||||||||||||||||||
|
||||||||||||||||||||||||||||||
Parameter histogram for exponential model (uncertainty of the parameter)
Exponential model plot, with confidence bounds around optimized model
| ||||||||||||||||||||||
| Model | Deviance | Δ | DF | χ20.95,df | χ20.95,1 | Good fit? | Parameters | LD50 |
|---|---|---|---|---|---|---|---|---|
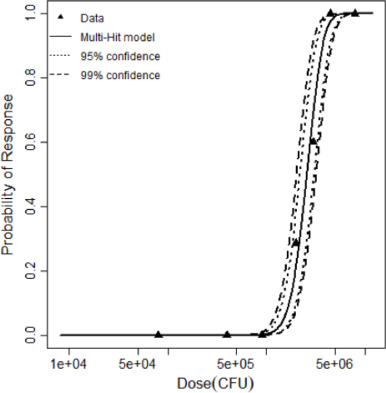
| Multi-hit | 1.09 | 15.69 | 5 | 11.1 | 3.84 | Yes | k = 4.12 × 10−6 kmin=11 | 2,588,047 |