| Experiment ID |
1
|
|---|---|
| Pathogen | |
| Agent Strain |
CO92
|
| Contains Preferred Model |
Yes
|
| Reference |
Lathem, W. W., Crosby, S. D., Miller, L. . , V, & Goldman, W. E. (2005). Progression of primary pneumonic plague: A mouse model of infection, pathology, and bacterial transcriptional activity. Proceedings of the National Academy of Sciences of the United States of America, 102, 17786-17791. https://doi.org/10.1073/pnas.0506840102 |
| Exposure Route |
intranasal
|
| Response |
death
|
| Host type |
mice
|
||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # of Doses |
4.00
|
||||||||||||||||||||
| Dose Units |
CFU
|
||||||||||||||||||||
| Μodel |
exponential
|
||||||||||||||||||||
| k |
1.63E-03
|
||||||||||||||||||||
| LD50/ID50 |
4.26E+02
|
||||||||||||||||||||
| Experiment Dataset |
|
Description
|
| ||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||
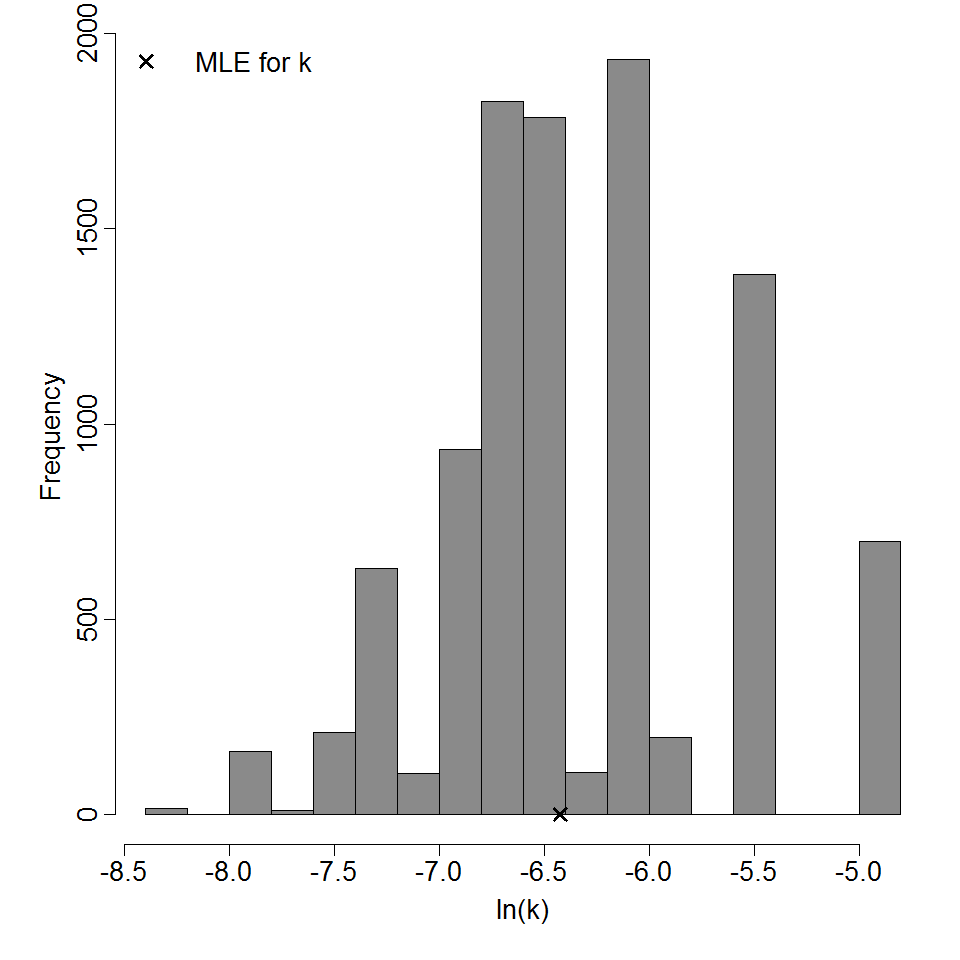
Parameter histogram for exponential model (uncertainty of the parameter)
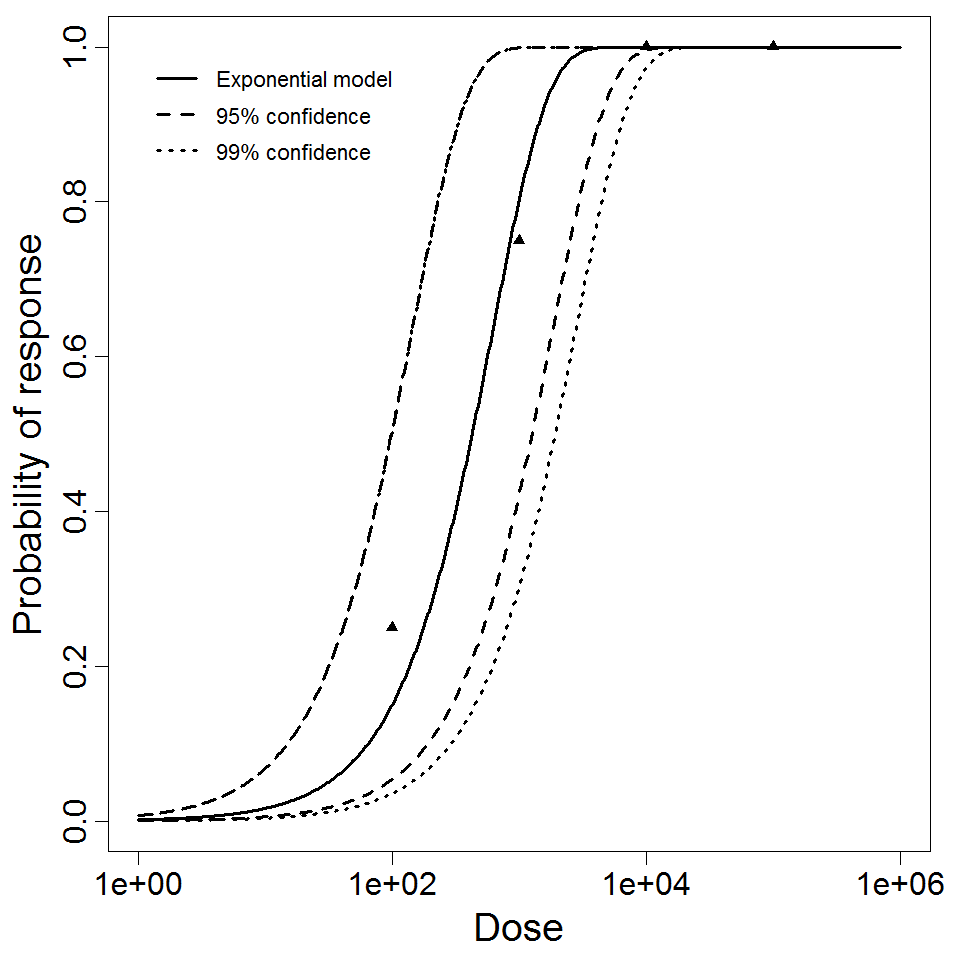
Exponential model plot, with confidence bounds around optimized model