Description
|
| ||||||||||||||||||||||
| ||||||||||||||||||||||||||||||
# of Doses
8.00
Μodel
LD50/ID50
6.17E+00
Dose Units
Exposure Route
Contains Preferred Model
a
2.53E-02
k
6.17E+00
Experiment ID
70
Host type
Experiment Dataset
Description
|
| ||||||||||||||||||||||
| ||||||||||||||||||||||||||||||
# of Doses
8.00
Μodel
N50
96.1
LD50/ID50
96.1
Dose Units
Response
Exposure Route
Contains Preferred Model
a
9.6E-2
Agent Strain
CJN strain (unpassaged
Experiment ID
125
Host type
Experiment Dataset
Description
|
|
||||||||||||||||||||||
|
||||||||||||||||||||||||||||||
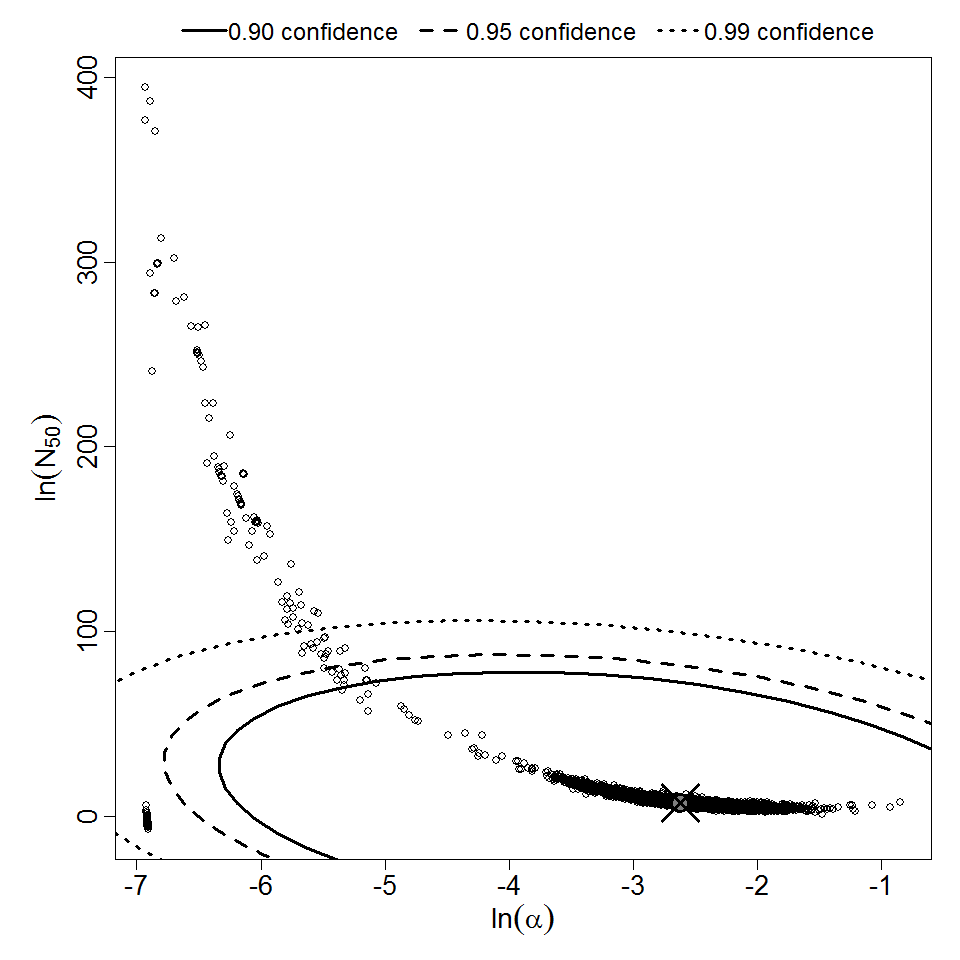
Parameter scatter plot for beta Poisson model ellipses signify the 0.9, 0.95 and 0.99 confidence of the parameters.
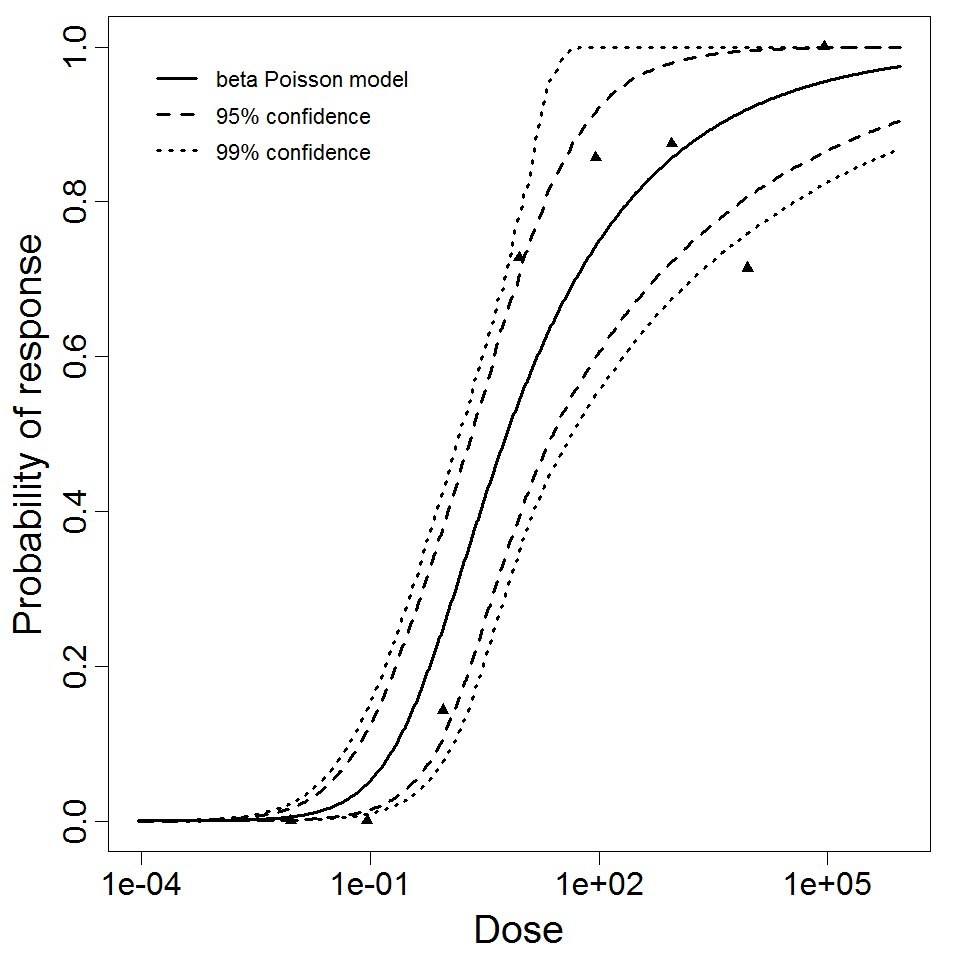
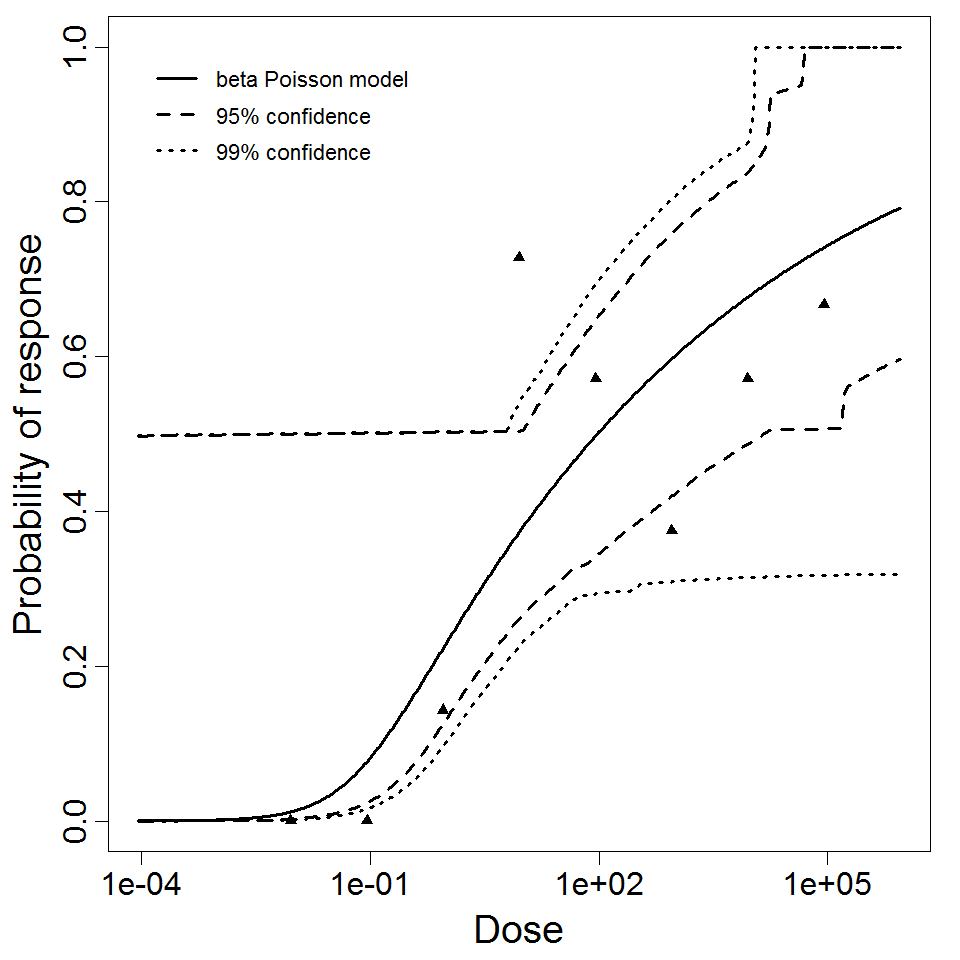
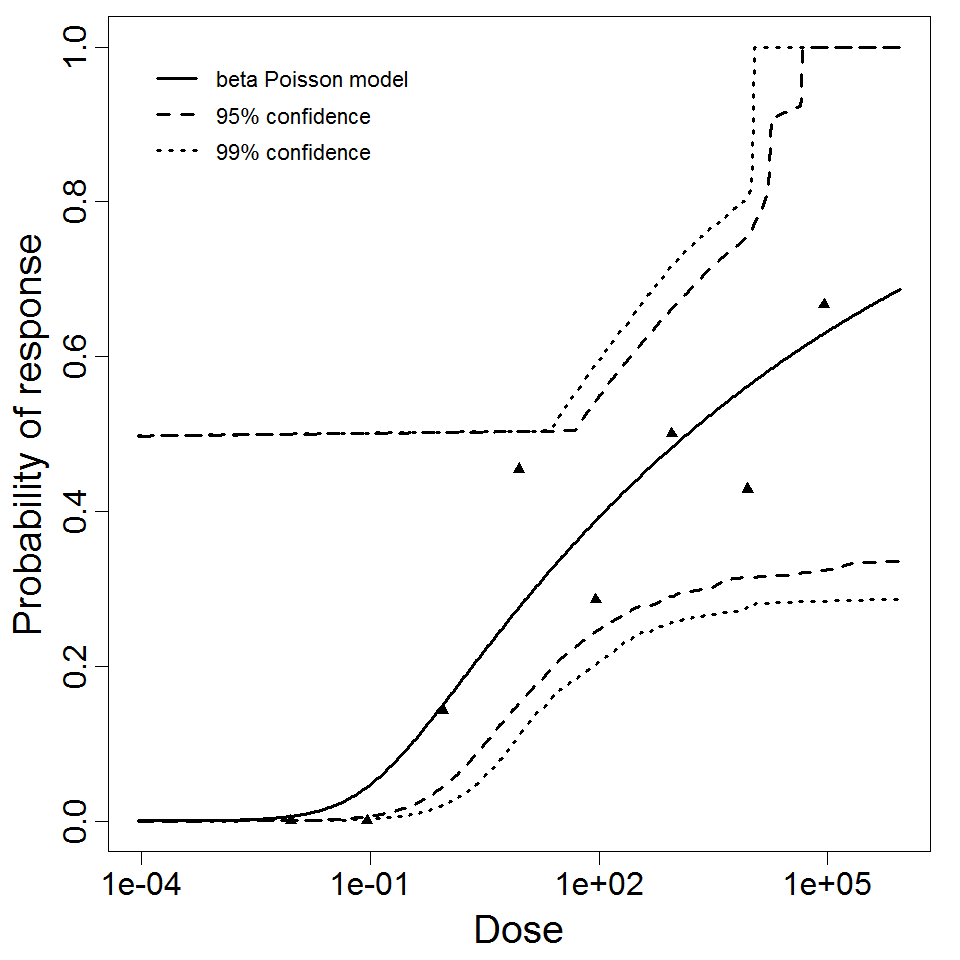
beta Poisson model plot, with confidence bounds around optimized model
# of Doses
8.00
Μodel
N50
1.47E+03
LD50/ID50
1.47E+03
Dose Units
Response
Exposure Route
Contains Preferred Model
a
7.28E-02
Agent Strain
CJN strain (unpassaged)
Experiment ID
71
Host type