Description
|
|
||||||||||||||||||||||
|
||||||||||||||||||||||||||||||||||||||

Parameter histogram for exponential model (uncertainty of the parameter)

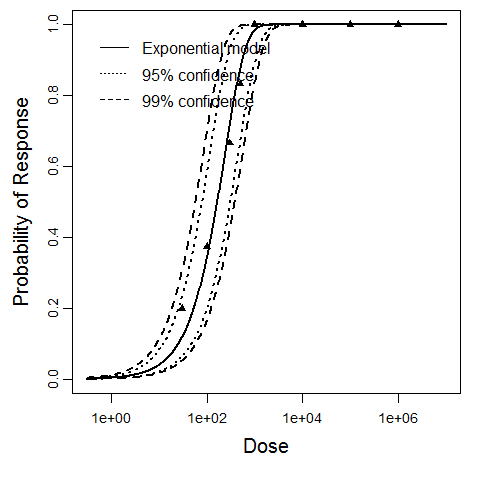
Exponential model plot, with confidence bounds around optimized model
# of Doses
8.00
Μodel
LD50/ID50
1.32E+02
Dose Units
Response
Exposure Route
Contains Preferred Model
k
5.26E-03
Agent Strain
Iowa isolate
Experiment ID
139
Host type
Description
|
|
||||||||||||||||||||||
|
||||||||||||||||||||||||||||||
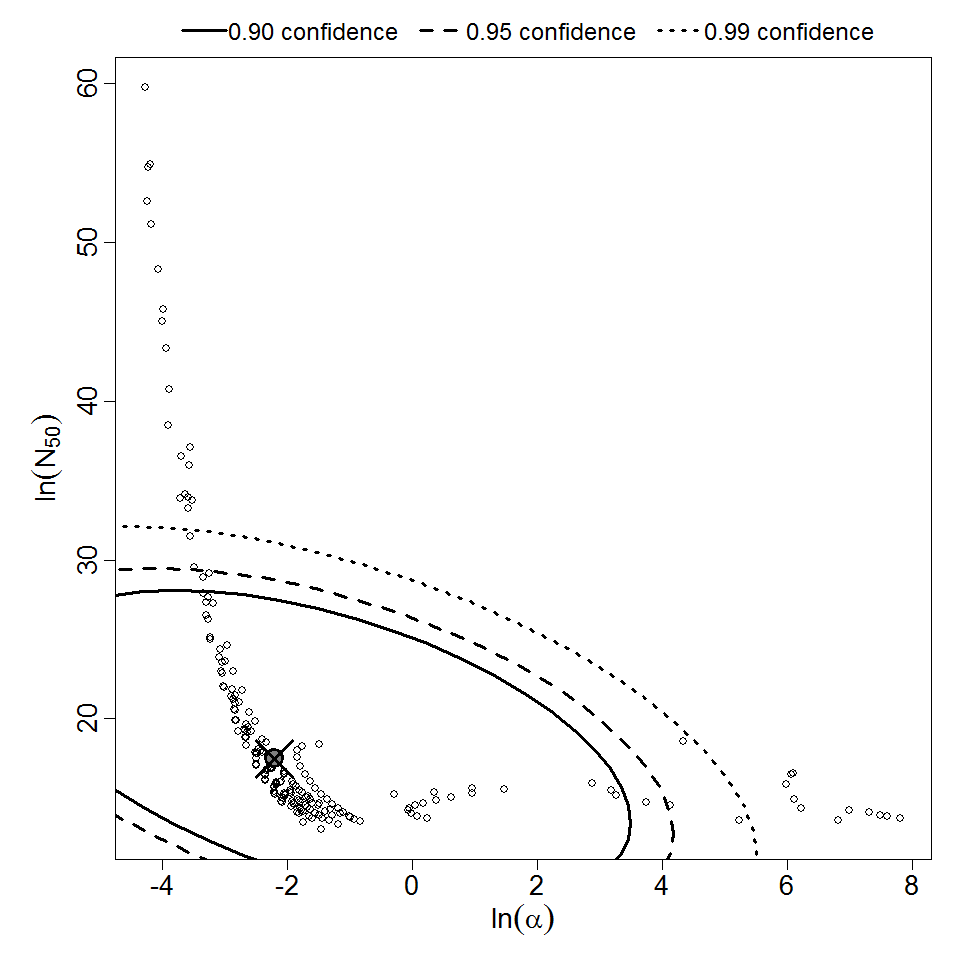
Parameter scatter plot for beta Poisson model ellipses signify the 0.9, 0.95 and 0.99 confidence of the parameters.

beta Poisson model plot, with confidence bounds around optimized model
# of Doses
6.00
Μodel
N50
3.88E+07
LD50/ID50
3.88E+07
Dose Units
Response
Exposure Route
Contains Preferred Model
a
1.10E-01
Agent Strain
Inaba 569B
Experiment ID
128
Host type
Description
|
|
||||||||||||||||||||||
|
||||||||||||||||||||||||||||||
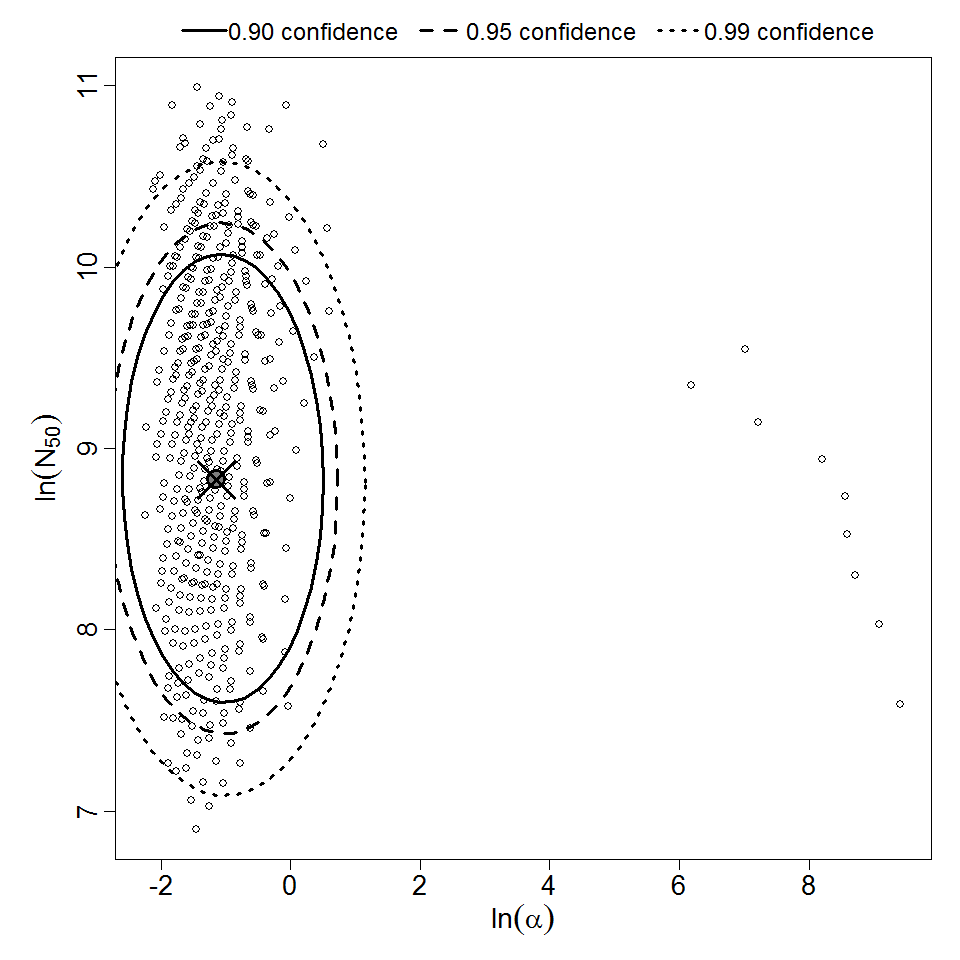
Parameter scatter plot for beta Poisson model ellipses signify the 0.9, 0.95 and 0.99 confidence of the parameters.

beta Poisson model plot, with confidence bounds around optimized model
# of Doses
6.00
Μodel
N50
6.82E+03
LD50/ID50
6.82E+03
Dose Units
Response
Exposure Route
Contains Preferred Model
a
3.18E-01
Agent Strain
Inaba 569B
Experiment ID
126
Host type
Description
|
|
||||||||||||||||||||||
|
||||||||||||||||||||||||||||||
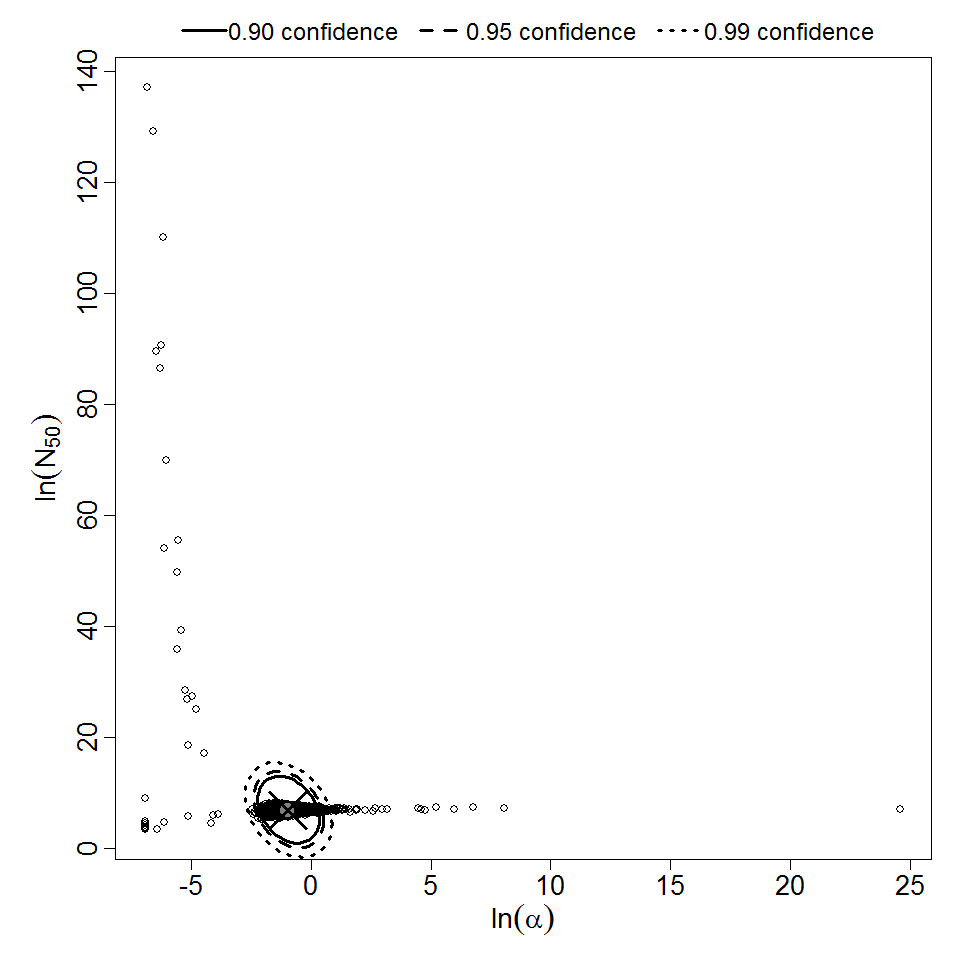
Parameter scatter plot for beta Poisson model ellipses signify the 0.9, 0.95 and 0.99 confidence of the parameters.
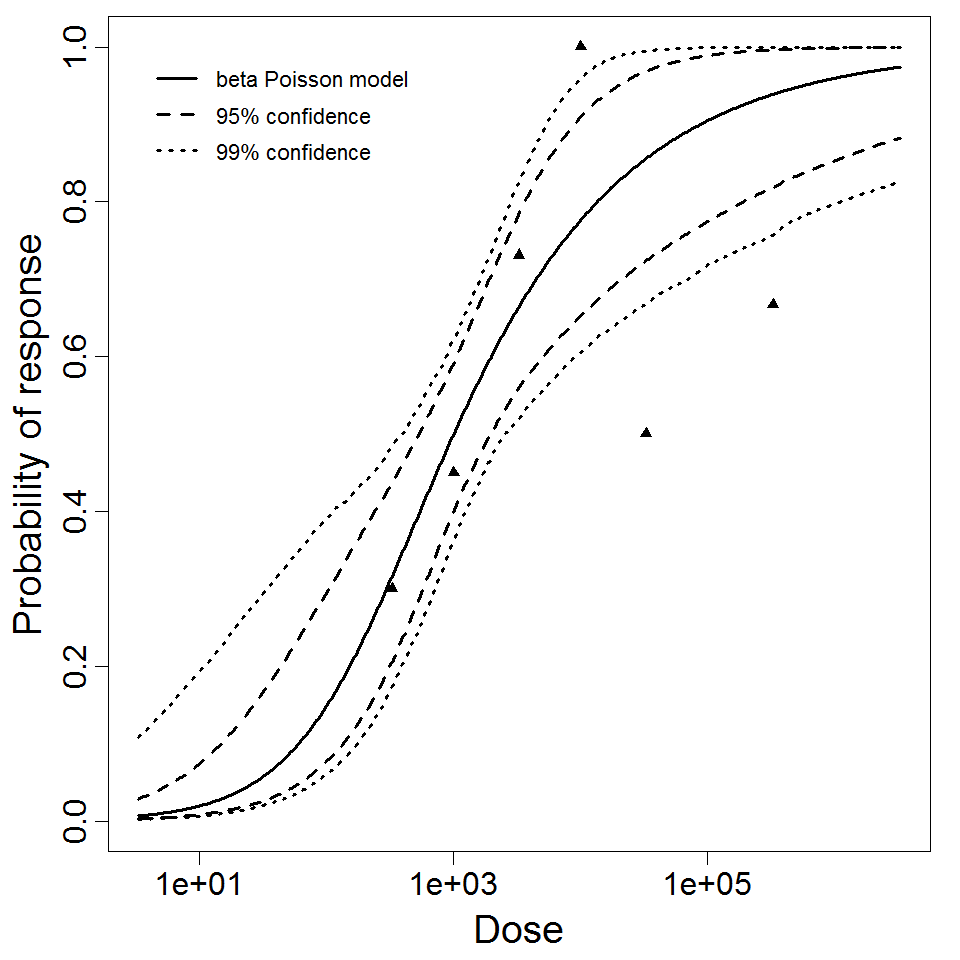
beta Poisson model plot, with confidence bounds around optimized model
# of Doses
6.00
Μodel
N50
1.01E+03
LD50/ID50
1.01E+03
Dose Units
Response
Exposure Route
Contains Preferred Model
a
3.74E-01
Agent Strain
strain 12
Experiment ID
112
Host type
Description
|
| ||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||
# of Doses
8.00
Μodel
LD50/ID50
1.65E+02
Dose Units
Response
Exposure Route
Contains Preferred Model
k
4.19E-03
Agent Strain
Iowa strain
Experiment ID
108
Host type
Experiment Dataset
Pagination
- Previous page
- Page 7