Description
|
| ||||||||||||||||||||||
| ||||||||||||||||||||||||||||||
# of Doses
8.00
Μodel
N50
6.85E+07
LD50/ID50
6.85E+07
Dose Units
Response
Exposure Route
Contains Preferred Model
a
2.21E-01
Agent Strain
EPEC B171-8 (serotype O11:NM)
Experiment ID
214, 216, 217
Host type
Experiment Dataset
Description
|
|
||||||||||||||||||||||
|
||||||||||||||||||||||||||||||

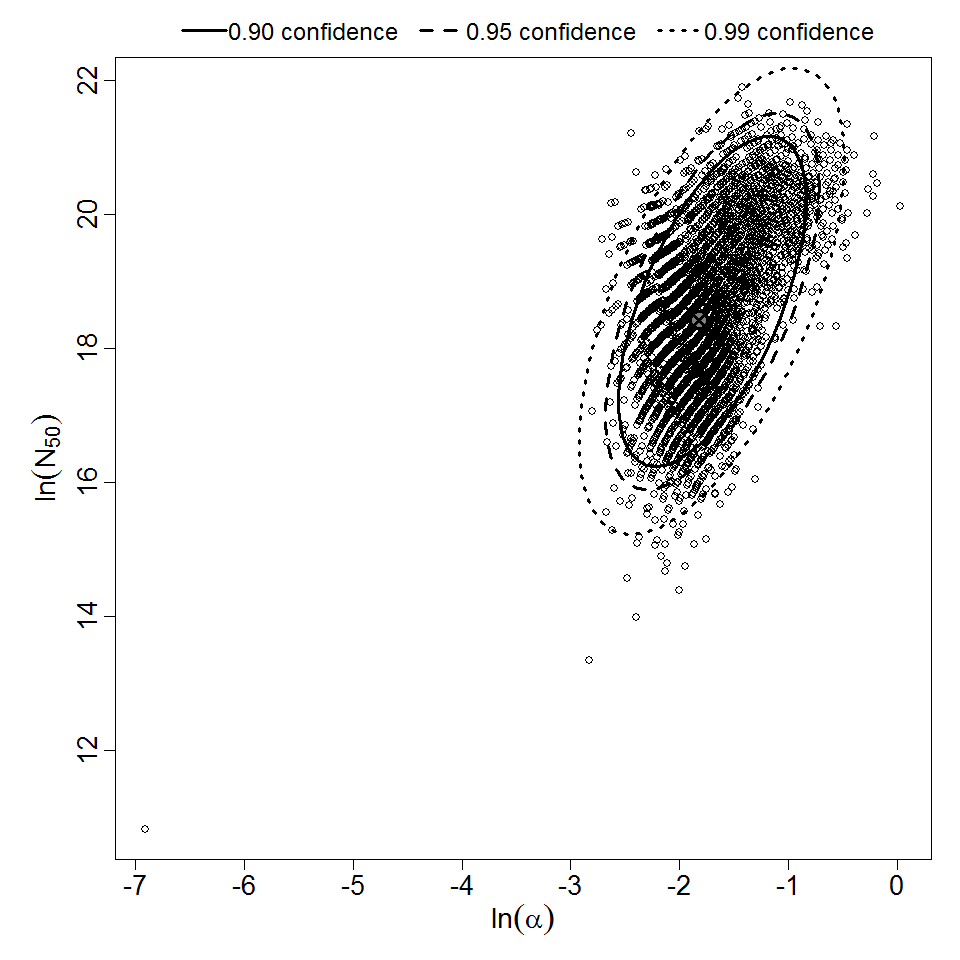
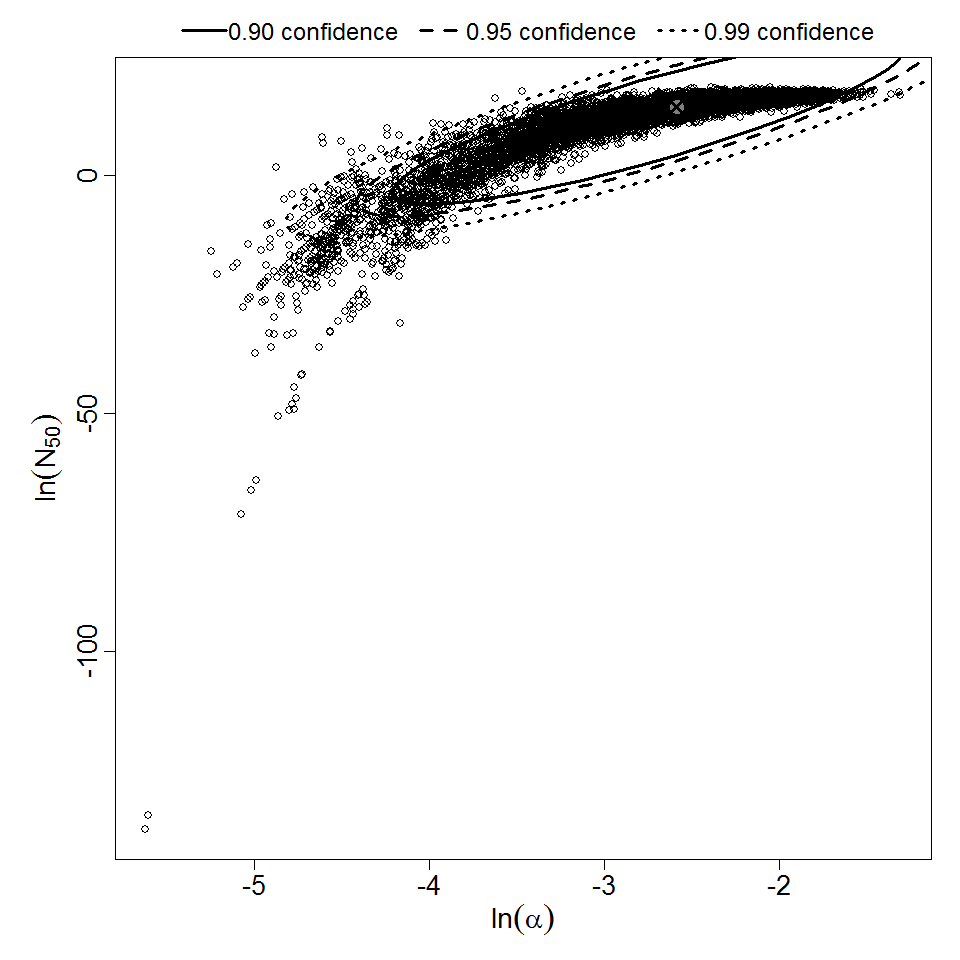
Parameter scatter plot for beta Poisson model ellipses signify the 0.9, 0.95 and 0.99 confidence of the parameters.

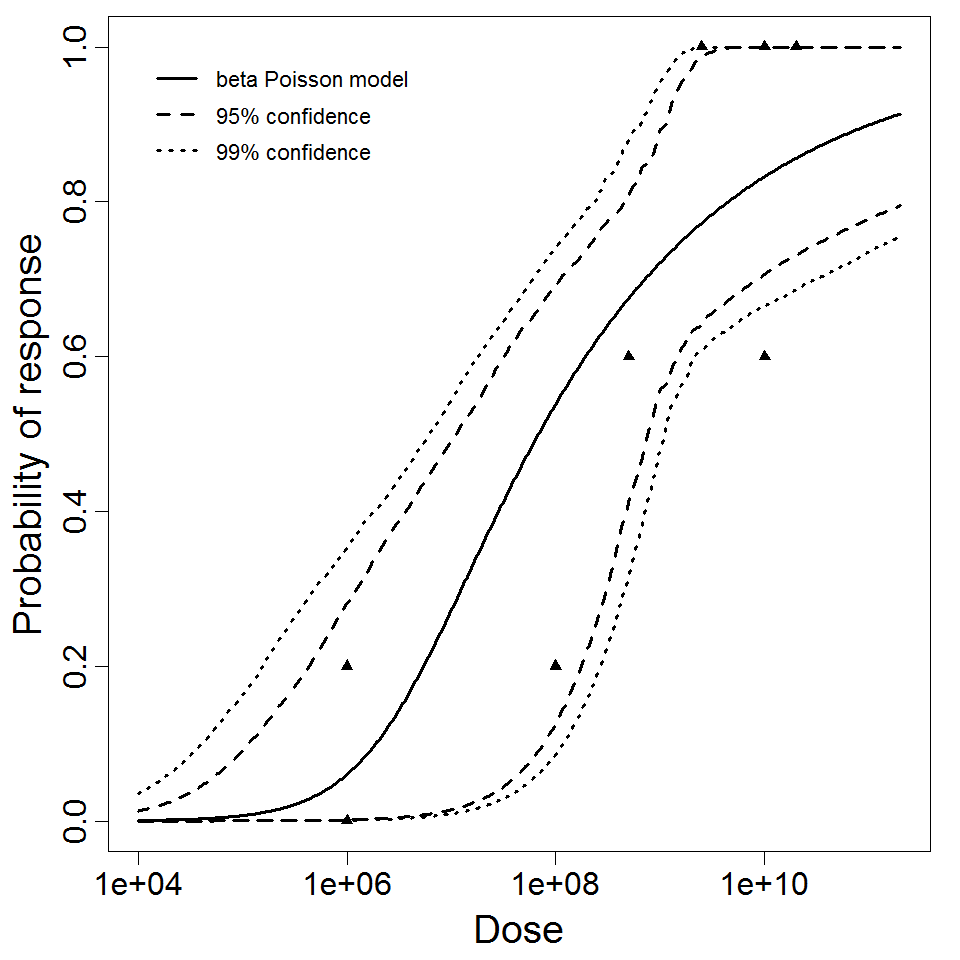
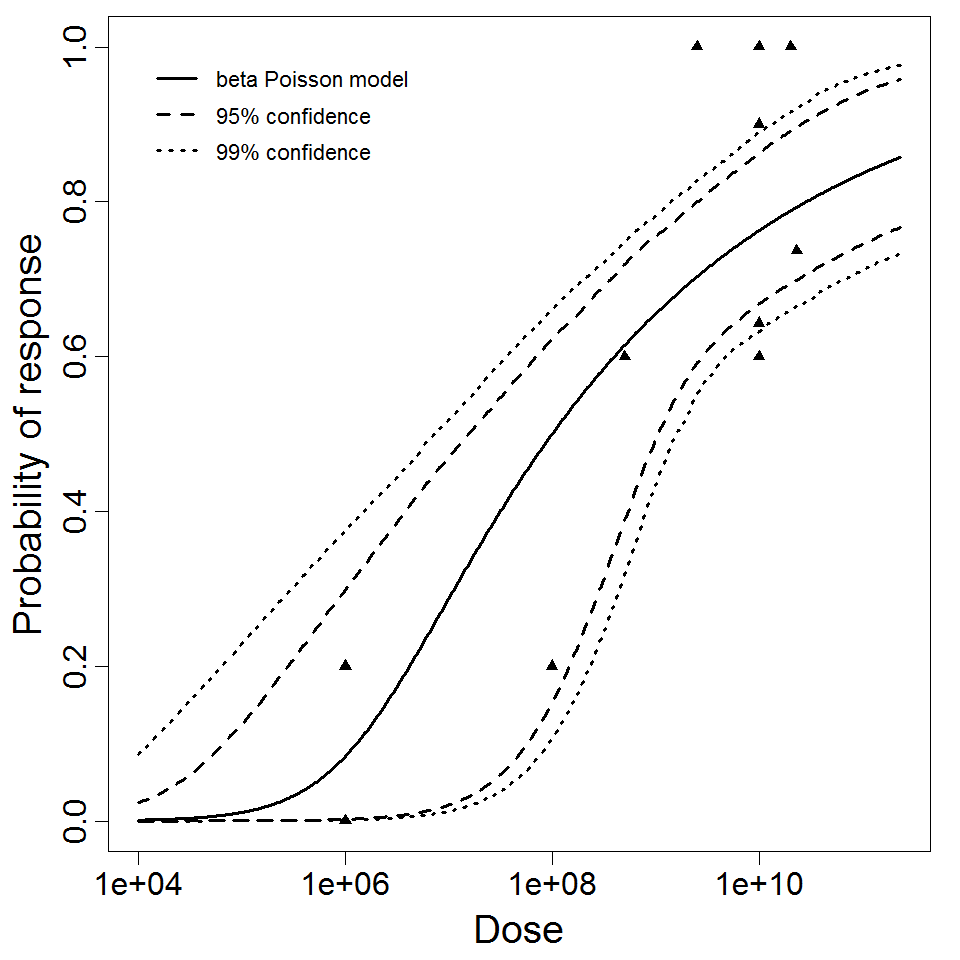
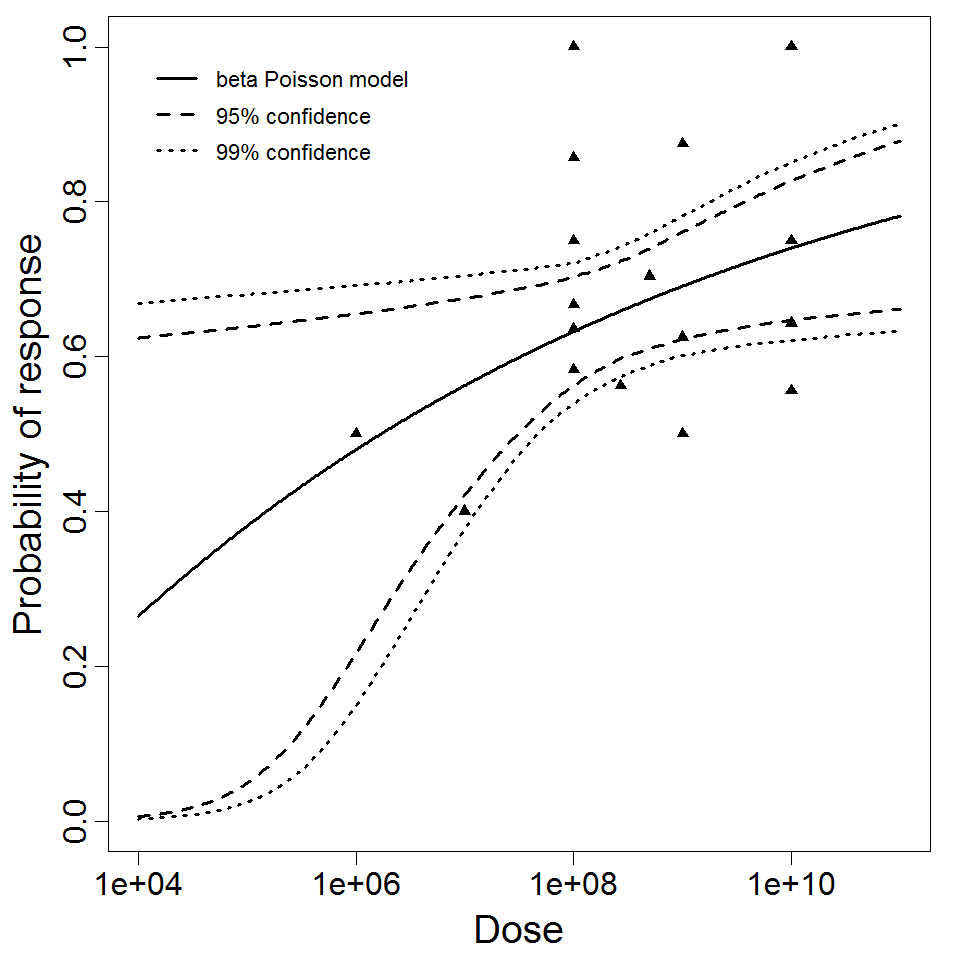
beta Poisson model plot, with confidence bounds around optimized model.
# of Doses
4.00
Μodel
N50
1.68E+01
LD50/ID50
1.68E+01
Dose Units
Response
Exposure Route
Contains Preferred Model
a
2.7E-01
Agent Strain
*C. hominis*, TU502
Experiment ID
181
Host type
Description
|
| ||||||||||||||||||||||
| ||||||||||||||||||||||||||||||
# of Doses
11.00
Μodel
N50
9.98E+07
LD50/ID50
9.98E+07
Dose Units
Response
Exposure Route
Contains Preferred Model
a
1.62E-01
Agent Strain
EPEC E2348/69 (O127:H6)
Experiment ID
153, 157, 159, 214, 216, 217
Host type
Experiment Dataset
Description
|
| ||||||||||||||||||||||
| ||||||||||||||||||||||||||||||
# of Doses
19.00
Μodel
N50
1.7E+06
LD50/ID50
1.7E+06
Dose Units
Response
Exposure Route
Contains Preferred Model
a
7.54E-02
Agent Strain
ETEC B7A
Experiment ID
142, 143, 144, 145, 147, 151, 161, 162, 163, 164, 168, 169, 170, 172
Host type
Experiment Dataset