Description
|
| ||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||
# of Doses
5.00
Μodel
LD50/ID50
6.63E+01
Dose Units
Response
Exposure Route
Contains Preferred Model
k
1.04E-02
Agent Strain
KHW
Experiment ID
17
Host type
Experiment Dataset
Description
|
|
||||||||||||||||||||||
|
||||||||||||||||||||||||||||||
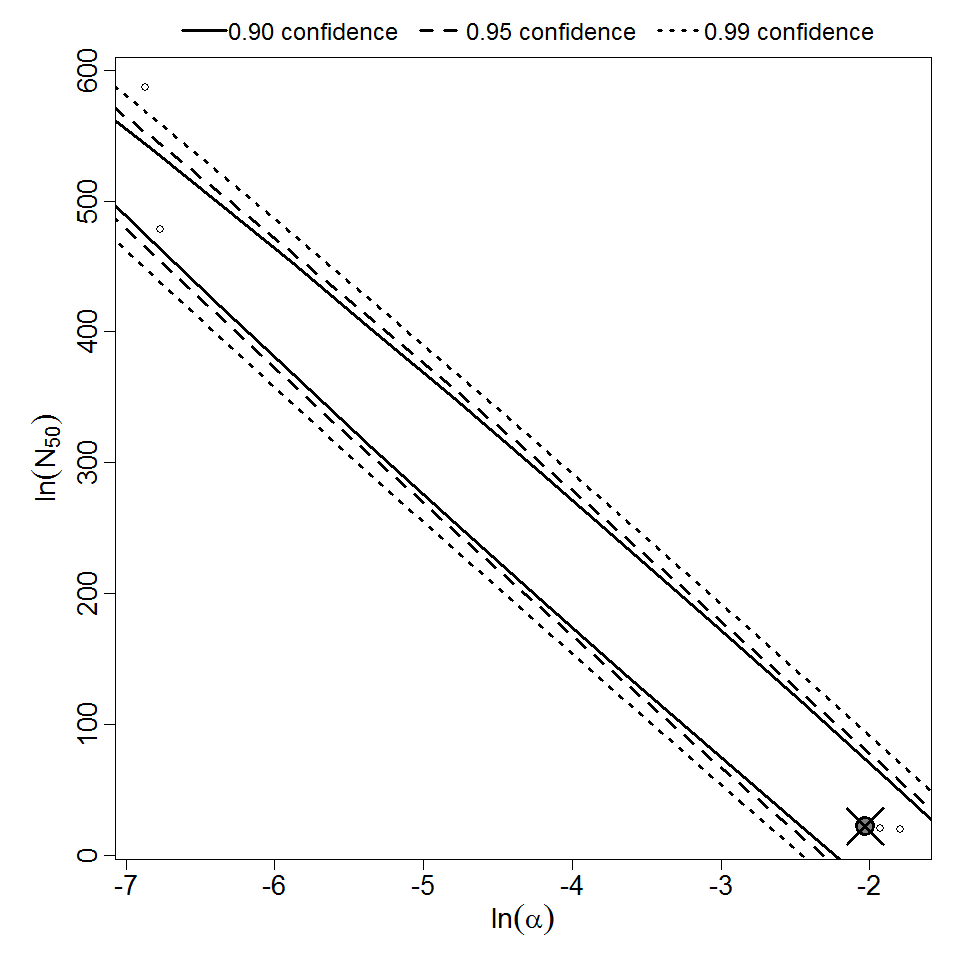
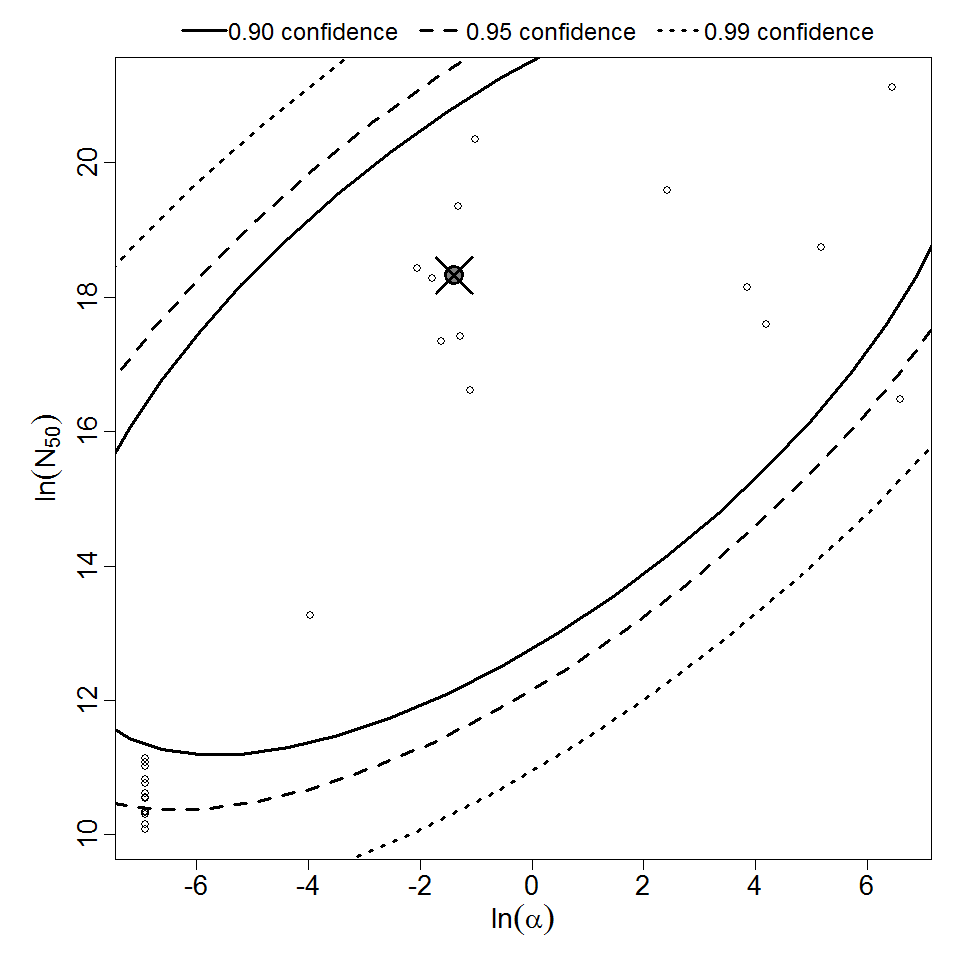
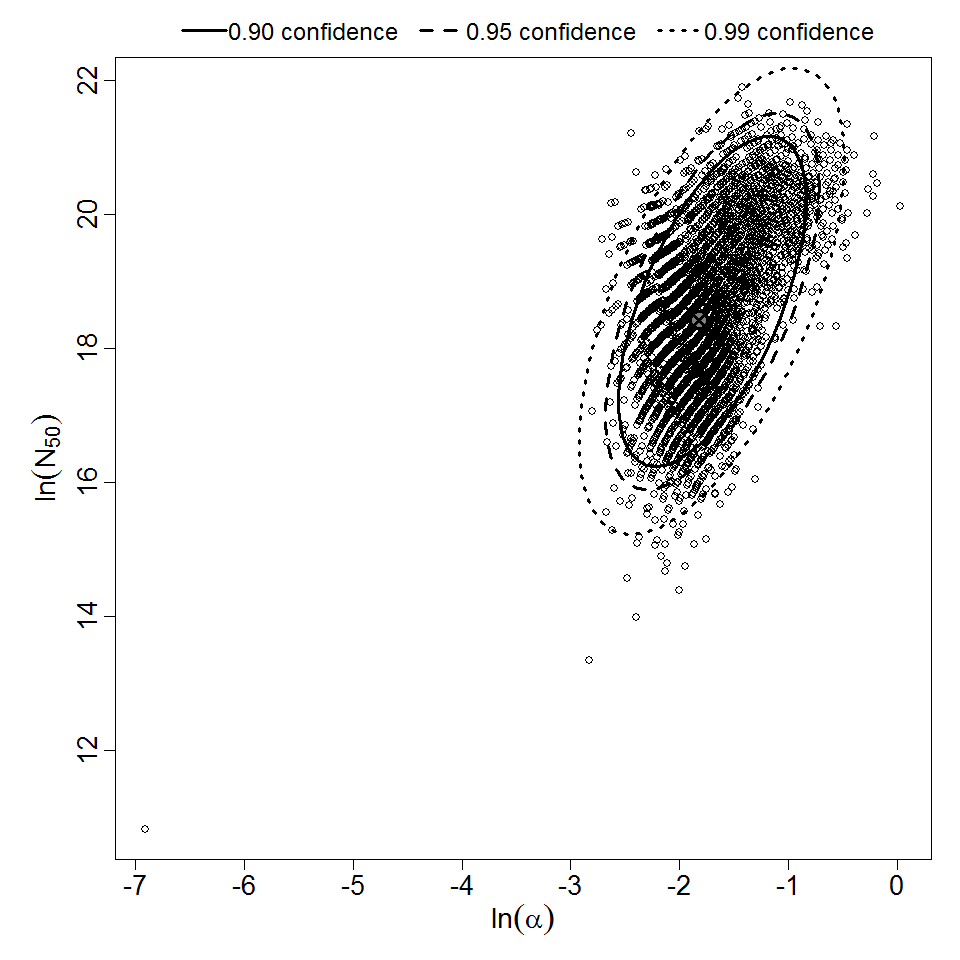
Parameter scatter plot for beta Poisson model ellipses signify the 0.9, 0.95 and 0.99 confidence of the parameters.

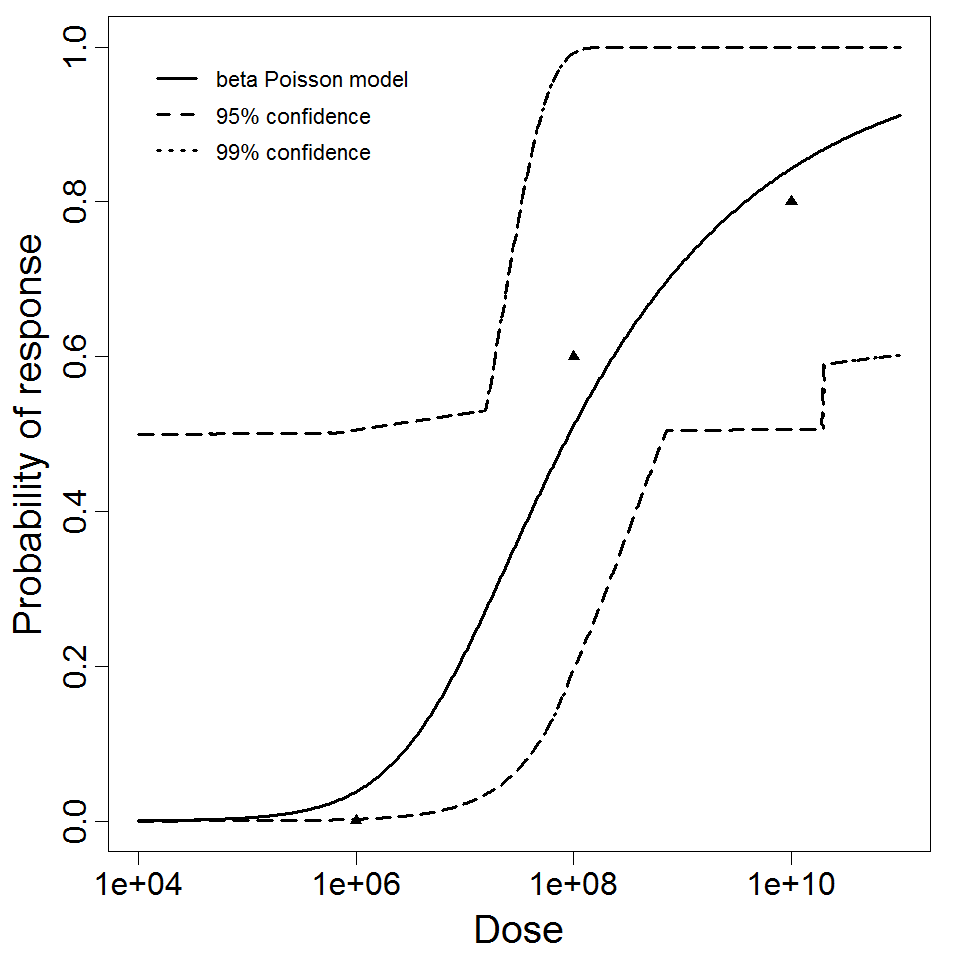
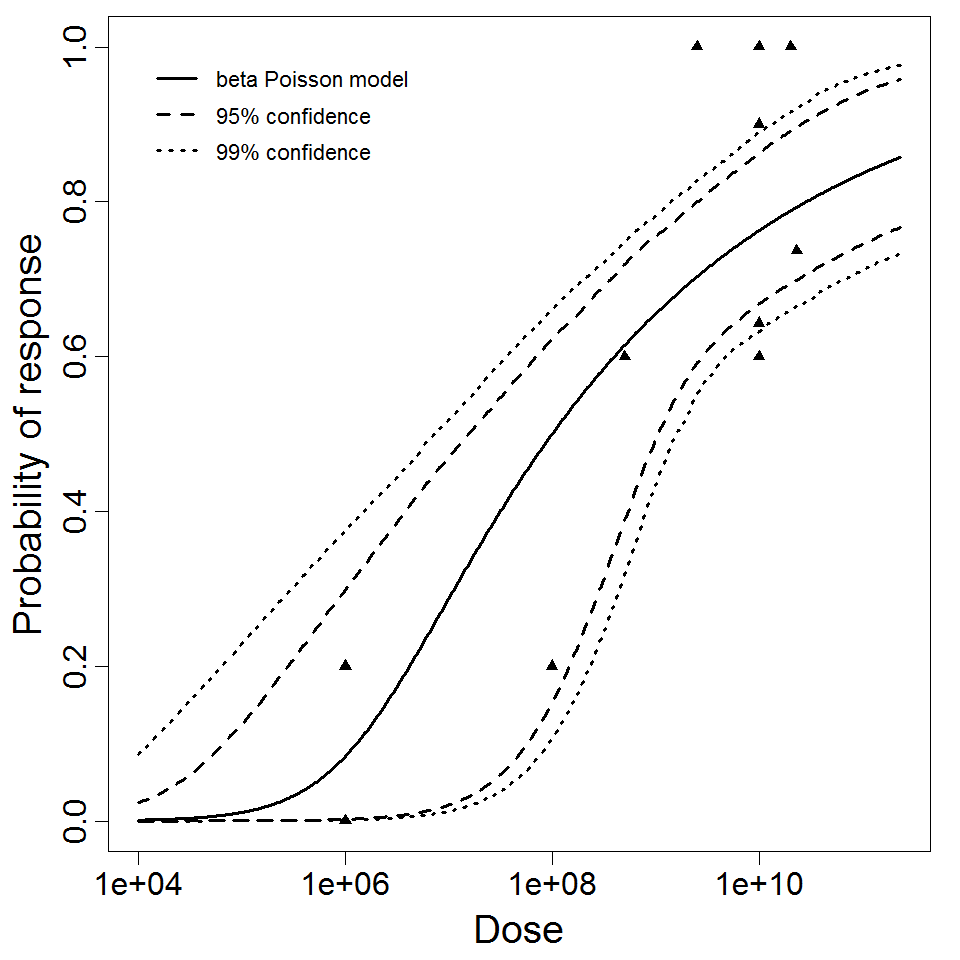
beta Poisson model plot, with confidence bounds around optimized model
# of Doses
7.00
Μodel
N50
2.91E+09
LD50/ID50
2.91E+09
Dose Units
Response
Exposure Route
Contains Preferred Model
a
1.31E-01
Agent Strain
Inaba 569B (classical)
Experiment ID
167
Host type
Description
|
| ||||||||||||||||||||||
| ||||||||||||||||||||||||||||||
# of Doses
3.00
Μodel
N50
9.1E+07
LD50/ID50
9.1E+07
Dose Units
Response
Exposure Route
Contains Preferred Model
a
2.5E-01
Agent Strain
ETEC 214-4 (ST)
Experiment ID
165
Host type
Experiment Dataset
Description
|
| ||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||
# of Doses
13.00
Μodel
LD50/ID50
3.56E+05
Dose Units
Response
Exposure Route
Contains Preferred Model
k
1.95E-06
Agent Strain
EPEC E2348/69 (O127:H6)
Experiment ID
154, 156, 158, 160, 219, 220, 221
Host type
Experiment Dataset
Description
|
| ||||||||||||||||||||||
| ||||||||||||||||||||||||||||||
# of Doses
11.00
Μodel
N50
9.98E+07
LD50/ID50
9.98E+07
Dose Units
Response
Exposure Route
Contains Preferred Model
a
1.62E-01
Agent Strain
EPEC E2348/69 (O127:H6)
Experiment ID
153, 157, 159, 214, 216, 217
Host type
Experiment Dataset
Description
|
| ||||||||||||||||||||||
| ||||||||||||||||||||||||||||||
# of Doses
19.00
Μodel
N50
1.7E+06
LD50/ID50
1.7E+06
Dose Units
Response
Exposure Route
Contains Preferred Model
a
7.54E-02
Agent Strain
ETEC B7A
Experiment ID
142, 143, 144, 145, 147, 151, 161, 162, 163, 164, 168, 169, 170, 172
Host type
Experiment Dataset
Description
|
|
||||||||||||||||||||||
|
||||||||||||||||||||||||||||||
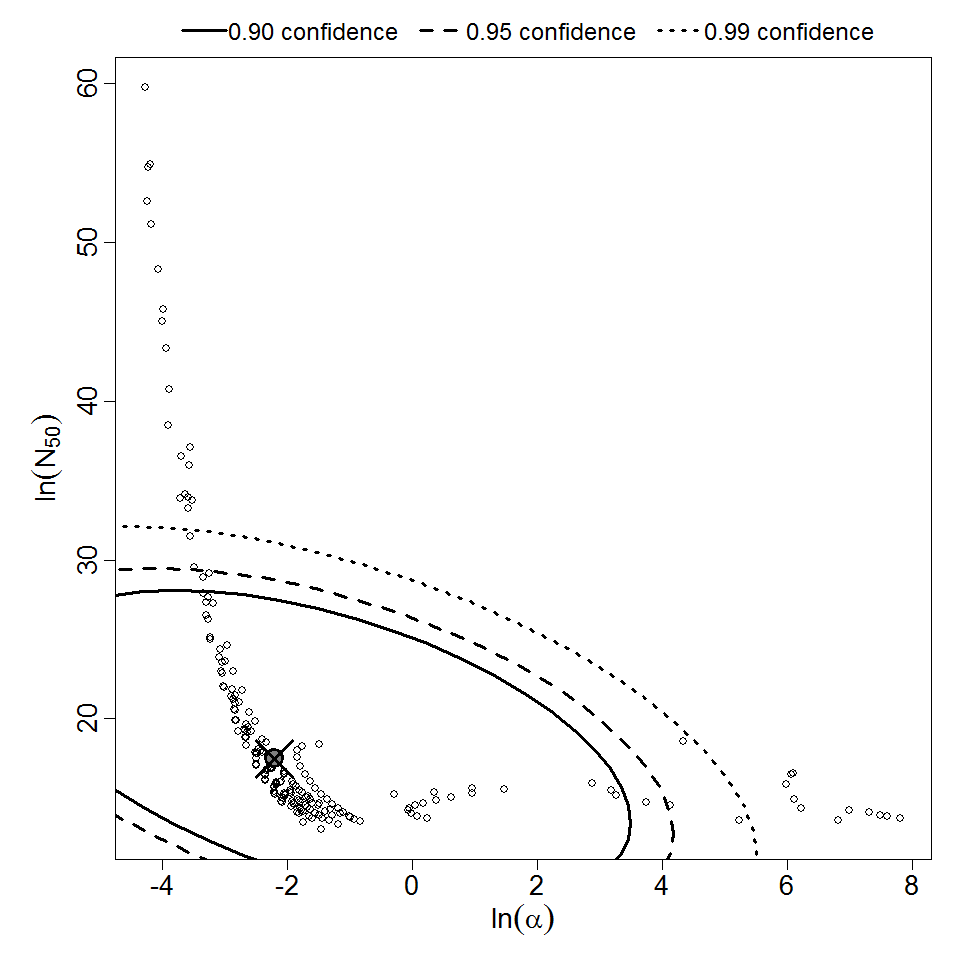
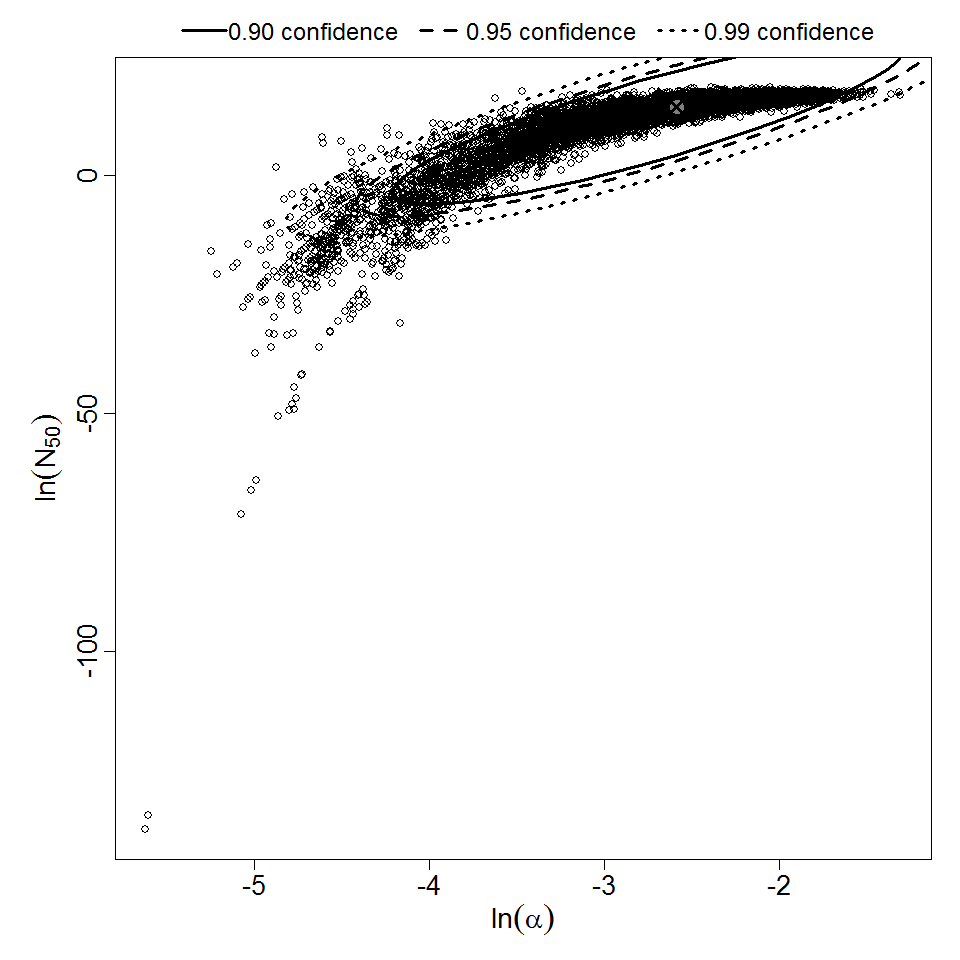
Parameter scatter plot for beta Poisson model ellipses signify the 0.9, 0.95 and 0.99 confidence of the parameters.

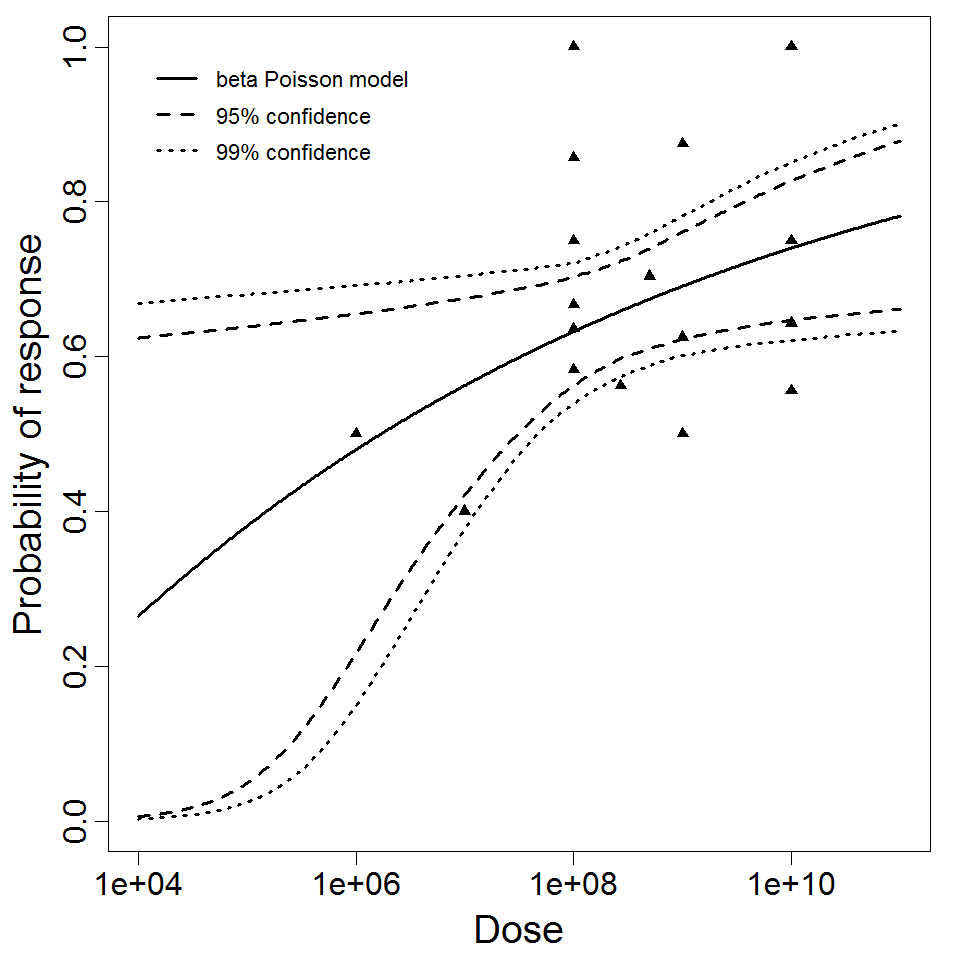
beta Poisson model plot, with confidence bounds around optimized model
# of Doses
6.00
Μodel
N50
3.88E+07
LD50/ID50
3.88E+07
Dose Units
Response
Exposure Route
Contains Preferred Model
a
1.10E-01
Agent Strain
Inaba 569B
Experiment ID
128
Host type
Description
|
|
||||||||||||||||||||||
|
||||||||||||||||||||||||||||||
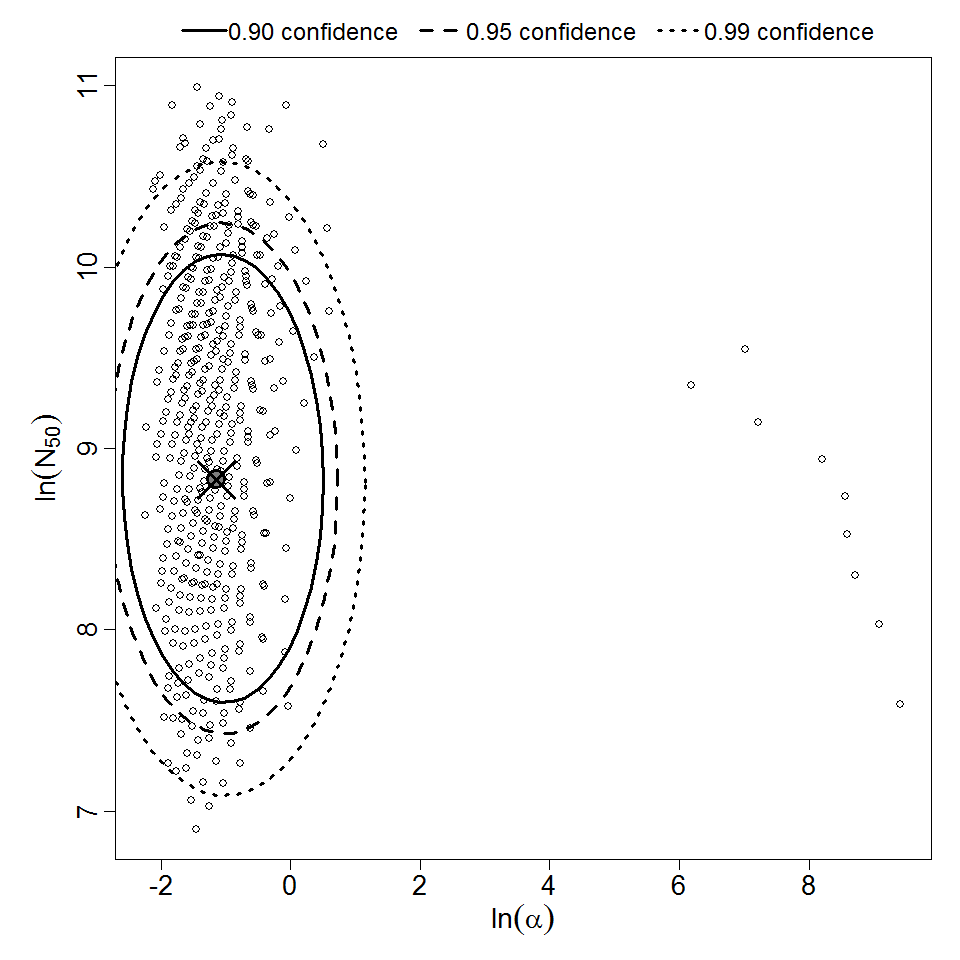
Parameter scatter plot for beta Poisson model ellipses signify the 0.9, 0.95 and 0.99 confidence of the parameters.

beta Poisson model plot, with confidence bounds around optimized model
# of Doses
6.00
Μodel
N50
6.82E+03
LD50/ID50
6.82E+03
Dose Units
Response
Exposure Route
Contains Preferred Model
a
3.18E-01
Agent Strain
Inaba 569B
Experiment ID
126
Host type
Description
|
| ||||||||||||||||||||||
| ||||||||||||||||||||||||||||||||||||||
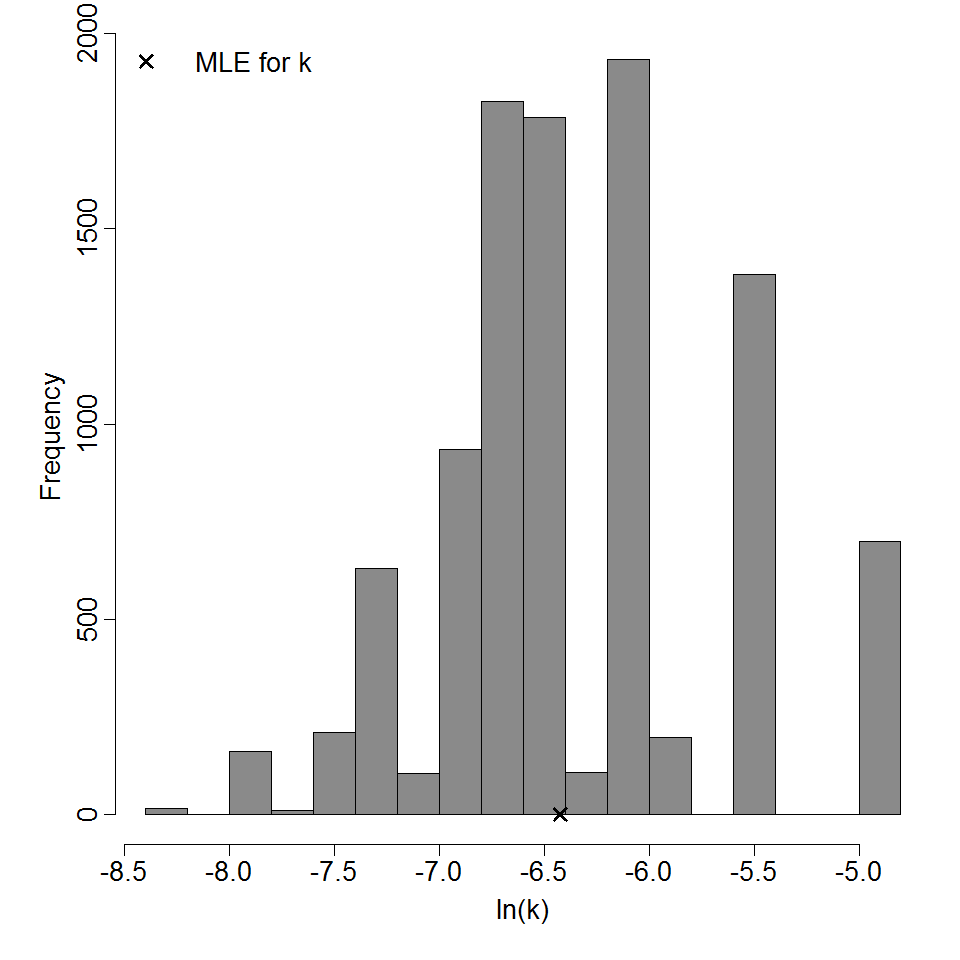
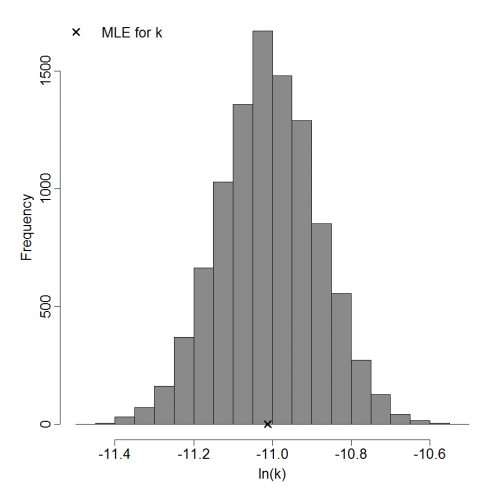
Parameter histogram for exponential model (uncertainty of the parameter)
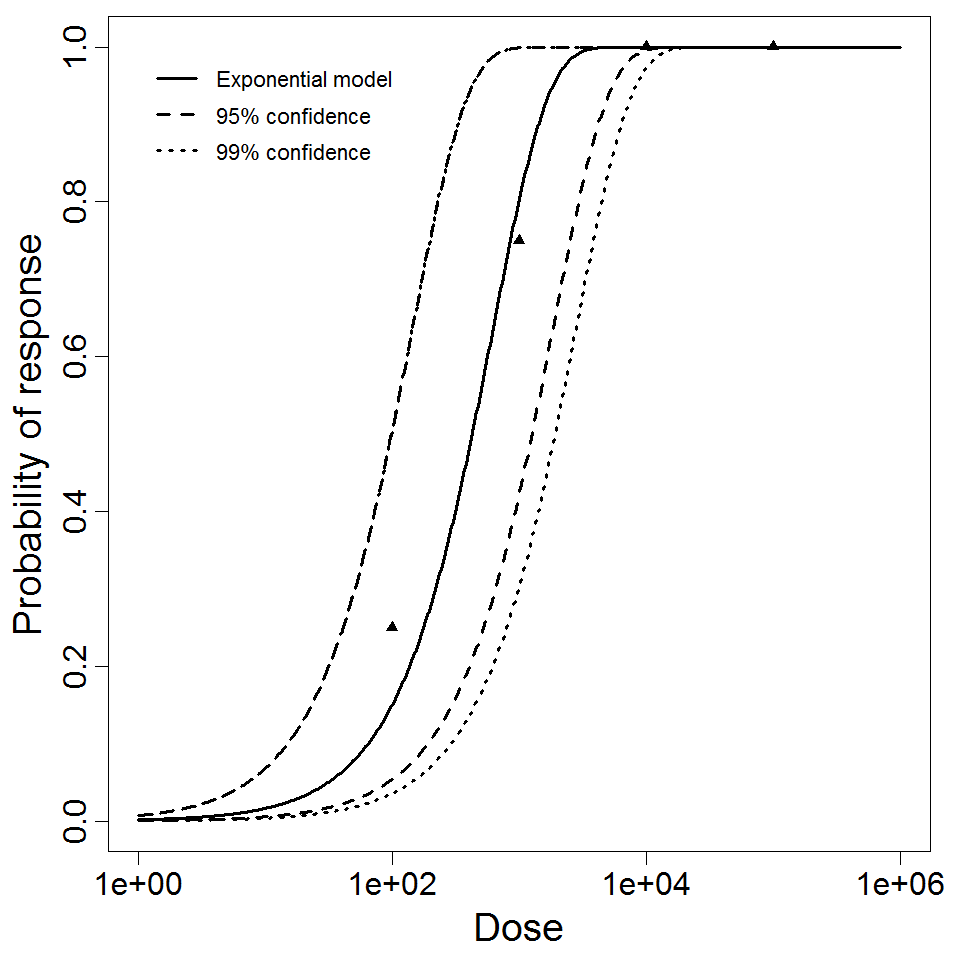
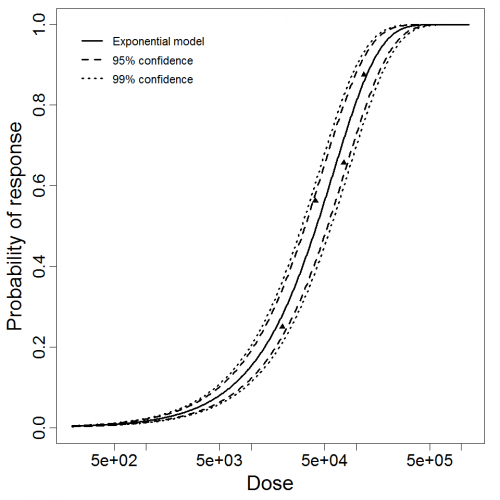
Exponential model plot, with confidence bounds around optimized model
# of Doses
4.00
Μodel
LD50/ID50
4.26E+02
Dose Units
Response
Exposure Route
Contains Preferred Model
k
1.63E-03
Agent Strain
CO92
Experiment ID
1
Host type
Experiment Dataset
| Dose | Dead | Survived | Total |
|---|---|---|---|
| 100 | 1 | 3 | 4 |
| 1000 | 3 | 1 | 4 |
| 1E+04 | 4 | 0 | 4 |
| 1E+05 | 4 | 0 | 4 |
Pagination
- Previous page
- Page 9